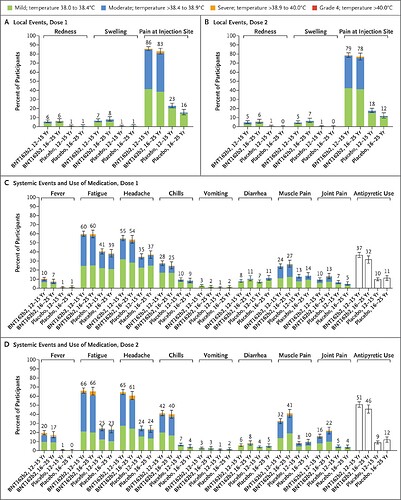
I agree with @nirgrahamuk about the tutorial. I will add some general advice about data layout.
- You must not mix characters and numeric values in a vector. That will force everything in the vector to be characters.
- Do not have column names with spaces. It can be done but it introduces needless complexity.
- Have each column of your data represent one thing. Instead of having a column that shows both the symptom (Fatigue or Pain) and whether the subject was in the vaccine or placebo cohort, have one column for the symptom and one for the cohort.
- Do not put data in the column headers. Instead of having a column for each study, make a column that stores the study name.
In the code below, I rearrange your data to satisfy those rules and then make a plot. The data wrangling will probably be rather confusing to a beginner. Focus on the difference between your original data and what I end up with.
Walter_et_al <- c(34, 31, 74, 31)
#names(Study1A) <-c('Study', 'Fatigue Vaccine', 'Fatigue Placebo', 'Inj site pain vaccine', 'Inj site pain Placebo')
Frenck_et_al <- c(60,41,86,23)
#names(Study2a)<-c('Study','Fatigue Vaccine', 'Fatigue Placebo', 'Inj site pain vaccine', 'Inj site pain Placbeo')
Ali_et_al <- c(47.9,36.6,93.1,34.8)
#names(Study3a)<-c('Study', 'Fatigue Vaccine', 'Fatigue Placebo', 'Inj site pain vaccine', 'Inj site pain Placbeo')
Category <- c('Fatigue Vaccine', 'Fatigue Placebo', 'Inj site pain Vaccine', 'Inj site pain Placebo')
DF <- data.frame(Category, Walter_et_al, Frenck_et_al, Ali_et_al)
DF
#> Category Walter_et_al Frenck_et_al Ali_et_al
#> 1 Fatigue Vaccine 34 60 47.9
#> 2 Fatigue Placebo 31 41 36.6
#> 3 Inj site pain Vaccine 74 86 93.1
#> 4 Inj site pain Placebo 31 23 34.8
library(tidyverse)
#> Warning: package 'tibble' was built under R version 4.1.2
DF <- DF |> mutate(Cohort = str_extract(Category, "[:alpha:]+$"),
Category = str_trim(str_remove(Category, "[:alpha:]+$")))
DF
#> Category Walter_et_al Frenck_et_al Ali_et_al Cohort
#> 1 Fatigue 34 60 47.9 Vaccine
#> 2 Fatigue 31 41 36.6 Placebo
#> 3 Inj site pain 74 86 93.1 Vaccine
#> 4 Inj site pain 31 23 34.8 Placebo
DFlong <- DF |> pivot_longer(Walter_et_al:Ali_et_al, names_to = "Study")
DFlong
#> # A tibble: 12 x 4
#> Category Cohort Study value
#> <chr> <chr> <chr> <dbl>
#> 1 Fatigue Vaccine Walter_et_al 34
#> 2 Fatigue Vaccine Frenck_et_al 60
#> 3 Fatigue Vaccine Ali_et_al 47.9
#> 4 Fatigue Placebo Walter_et_al 31
#> 5 Fatigue Placebo Frenck_et_al 41
#> 6 Fatigue Placebo Ali_et_al 36.6
#> 7 Inj site pain Vaccine Walter_et_al 74
#> 8 Inj site pain Vaccine Frenck_et_al 86
#> 9 Inj site pain Vaccine Ali_et_al 93.1
#> 10 Inj site pain Placebo Walter_et_al 31
#> 11 Inj site pain Placebo Frenck_et_al 23
#> 12 Inj site pain Placebo Ali_et_al 34.8
ggplot(DFlong, aes(x = Category, y = value, fill = Cohort)) +
geom_col(position = "dodge") + facet_wrap(~ Study, nrow = 1)
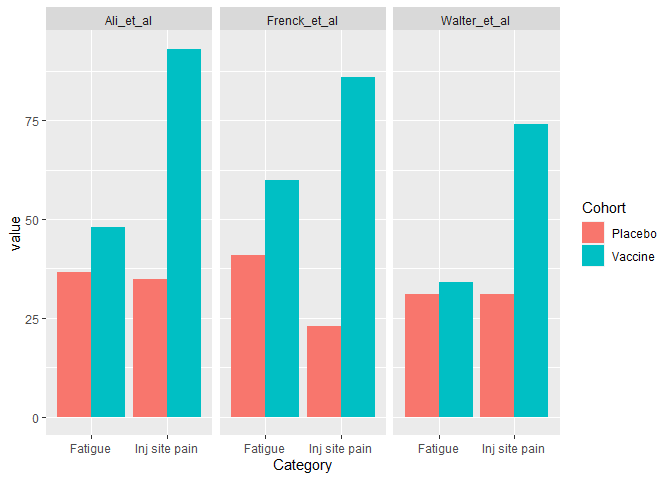
Created on 2022-05-10 by the reprex package (v2.0.1)