I need help to generate new date frame (from kss2.csv to kss2_v1.csv and vice vera)
Thanks a lot
KSS2.pdf (248.6 KB)
KSS2_v1.pdf (432.3 KB)
I need help to generate new date frame (from kss2.csv to kss2_v1.csv and vice vera)
Thanks a lot
KSS2.pdf (248.6 KB)
KSS2_v1.pdf (432.3 KB)
I suggest you keep some extra columns in the long form to make it easier to go back to the wide form.
library(tidyr)
library(dplyr)
DF <- data.frame(Nr_crt = 1:10,
AImin = round(rnorm(10),2),
BImin = round(rnorm(10),2),
AImax = round(rnorm(10),2),
BImax = round(rnorm(10),2),
AEmin = round(rnorm(10),2),
BEmin = round(rnorm(10),2),
AEmax = round(rnorm(10),2),
BEmax = round(rnorm(10),2))
DF
#> Nr_crt AImin BImin AImax BImax AEmin BEmin AEmax BEmax
#> 1 1 1.90 0.61 1.15 0.38 -0.02 0.71 -0.41 0.67
#> 2 2 -1.06 -1.55 -0.33 0.03 -2.20 -0.95 -0.02 -1.22
#> 3 3 1.60 0.00 0.66 0.06 -1.39 -0.97 0.16 0.79
#> 4 4 -1.33 -1.07 1.83 0.15 -0.04 -0.10 -0.83 -2.70
#> 5 5 0.17 -2.16 0.06 1.05 1.30 -0.69 -1.03 0.11
#> 6 6 -1.99 -0.69 2.14 -0.03 0.78 -0.14 -0.25 1.06
#> 7 7 0.63 1.52 -0.55 1.39 -0.21 -0.15 -1.51 0.21
#> 8 8 -0.07 -0.33 1.44 -0.47 1.03 -1.57 1.72 1.06
#> 9 9 -0.06 -0.05 -0.56 -0.75 0.75 0.48 1.15 0.78
#> 10 10 1.52 -0.21 -0.90 0.91 0.09 0.90 -0.53 -0.72
DFlong <- DF |> pivot_longer(AImin:BEmax) |>
mutate(CAP = substr(name, 1,1),
Tip = substr(name,2,2),
Poz = substr(name,3,5),
Nr_crt2 = row_number())
DFlong
#> # A tibble: 80 x 7
#> Nr_crt name value CAP Tip Poz Nr_crt2
#> <int> <chr> <dbl> <chr> <chr> <chr> <int>
#> 1 1 AImin 1.9 A I min 1
#> 2 1 BImin 0.61 B I min 2
#> 3 1 AImax 1.15 A I max 3
#> 4 1 BImax 0.38 B I max 4
#> 5 1 AEmin -0.02 A E min 5
#> 6 1 BEmin 0.71 B E min 6
#> 7 1 AEmax -0.41 A E max 7
#> 8 1 BEmax 0.67 B E max 8
#> 9 2 AImin -1.06 A I min 9
#> 10 2 BImin -1.55 B I min 10
#> # ... with 70 more rows
DF <- DFlong |> select(Nr_crt,name,value) |>
pivot_wider(names_from = "name", values_from = "value")
DF
#> # A tibble: 10 x 9
#> Nr_crt AImin BImin AImax BImax AEmin BEmin AEmax BEmax
#> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 1.9 0.61 1.15 0.38 -0.02 0.71 -0.41 0.67
#> 2 2 -1.06 -1.55 -0.33 0.03 -2.2 -0.95 -0.02 -1.22
#> 3 3 1.6 0 0.66 0.06 -1.39 -0.97 0.16 0.79
#> 4 4 -1.33 -1.07 1.83 0.15 -0.04 -0.1 -0.83 -2.7
#> 5 5 0.17 -2.16 0.06 1.05 1.3 -0.69 -1.03 0.11
#> 6 6 -1.99 -0.69 2.14 -0.03 0.78 -0.14 -0.25 1.06
#> 7 7 0.63 1.52 -0.55 1.39 -0.21 -0.15 -1.51 0.21
#> 8 8 -0.07 -0.33 1.44 -0.47 1.03 -1.57 1.72 1.06
#> 9 9 -0.06 -0.05 -0.56 -0.75 0.75 0.48 1.15 0.78
#> 10 10 1.52 -0.21 -0.9 0.91 0.09 0.9 -0.53 -0.72
Created on 2022-07-16 by the reprex package (v2.0.1)
Please take a look to pdf sttachrd..the new dataframe are not like pff attachef
I see that I exchanged the definitions of the Poz and Tip columns, so I fixed that. If you do not like the extra columns, you can choose the columns to keep, set their names, and reposition them with the select() function from the dplyr package.
DFlong <- DF |> pivot_longer(AImin:BEmax) |>
mutate(CAP = substr(name, 1,1),
Poz = substr(name,2,2),
Tip = substr(name,3,5),
Nr_crt2 = row_number())
DFlong2 <- DFlong |> select(Nr_crt = Nr_crt2, Val = value, CAP, Tip, Poz)
To get from DFlong2 back to DF, you will have to reconstruct the Nr_crt and name columns that are in DFlong. You can build name from CAP, Poz and Tip using the paste0() function.
See error below
Error in substr(name, 1, 1) : object 'name' not found
Here is my complete code that runs with no error.
library(tidyr)
library(dplyr)
DF <- data.frame(Nr_crt = 1:10,
AImin = round(rnorm(10),2),
BImin = round(rnorm(10),2),
AImax = round(rnorm(10),2),
BImax = round(rnorm(10),2),
AEmin = round(rnorm(10),2),
BEmin = round(rnorm(10),2),
AEmax = round(rnorm(10),2),
BEmax = round(rnorm(10),2))
DF
#> Nr_crt AImin BImin AImax BImax AEmin BEmin AEmax BEmax
#> 1 1 0.03 -0.58 -1.26 -0.28 1.51 0.32 -0.18 2.09
#> 2 2 1.35 -0.51 -0.64 -0.74 -0.35 0.47 2.38 0.04
#> 3 3 0.40 -1.23 0.29 -0.39 -0.63 0.03 0.10 0.54
#> 4 4 0.56 -0.74 0.71 -1.65 0.90 0.60 0.01 -1.11
#> 5 5 0.49 1.39 0.86 -0.45 -1.32 -1.26 0.17 -1.03
#> 6 6 -0.92 2.27 -1.31 1.65 -0.64 0.46 -0.37 1.28
#> 7 7 -0.43 0.06 -1.47 -0.54 -2.46 -1.16 0.03 -0.05
#> 8 8 0.14 0.59 0.58 -1.06 0.22 -0.06 0.82 -0.12
#> 9 9 -1.37 0.71 0.98 -0.47 -1.08 0.66 -0.02 -0.22
#> 10 10 -0.09 0.26 -1.26 0.61 1.41 0.60 -0.54 0.29
DFlong <- DF |> pivot_longer(AImin:BEmax) |>
mutate(CAP = substr(name, 1,1),
Poz = substr(name,2,2),
Tip = substr(name,3,5),
Nr_crt2 = row_number())
DFlong
#> # A tibble: 80 x 7
#> Nr_crt name value CAP Poz Tip Nr_crt2
#> <int> <chr> <dbl> <chr> <chr> <chr> <int>
#> 1 1 AImin 0.03 A I min 1
#> 2 1 BImin -0.58 B I min 2
#> 3 1 AImax -1.26 A I max 3
#> 4 1 BImax -0.28 B I max 4
#> 5 1 AEmin 1.51 A E min 5
#> 6 1 BEmin 0.32 B E min 6
#> 7 1 AEmax -0.18 A E max 7
#> 8 1 BEmax 2.09 B E max 8
#> 9 2 AImin 1.35 A I min 9
#> 10 2 BImin -0.51 B I min 10
#> # ... with 70 more rows
DFlong2 <- DFlong |> select(Nr_crt=Nr_crt2,Val=value,CAP,Tip,Poz)
DFlong2
#> # A tibble: 80 x 5
#> Nr_crt Val CAP Tip Poz
#> <int> <dbl> <chr> <chr> <chr>
#> 1 1 0.03 A min I
#> 2 2 -0.58 B min I
#> 3 3 -1.26 A max I
#> 4 4 -0.28 B max I
#> 5 5 1.51 A min E
#> 6 6 0.32 B min E
#> 7 7 -0.18 A max E
#> 8 8 2.09 B max E
#> 9 9 1.35 A min I
#> 10 10 -0.51 B min I
#> # ... with 70 more rows
Created on 2022-07-17 by the reprex package (v2.0.1)
If you still get an error, please show your data and all of the code leading up to the error.
[quote="FJCC, post:6, topic:142359"]
library(tidyr)
library(dplyr)
`Nr_crt Val CAP Tip Poz
1 0.31 A min I
2 0.34 A min I
3 0.37 A min I
4 0.3 A min I
5 0.28 A min I
6 0.31 A min I
7 0.34 A min I
8 0.35 A min I
9 0.32 A min I
10 0.35 A min I
11 0.27 A min I
12 0.29 A min I
13 0.31 A min I
14 0.28 A min I
15 0.3 A min I
16 0.29 A min I
17 0.29 A min I
18 0.3 A min I
19 0.32 A min I
20 0.29 A min I
21 0.33 A min I
22 0.3 A min I
23 0.31 A min I
24 0.32 A min I
25 0.33 A min I
26 0.31 A min I
27 0.29 A min I
28 0.31 A min I
29 0.32 A min I
30 0.28 A min I
31 0.32 A min I
32 0.33 A min I
33 0.33 A min I
34 0.31 A min I
35 0.32 A min I
36 0.31 A min I
37 0.3 A min I
38 0.3 A min I
39 0.32 A min I
40 0.32 A min I
41 0.32 A min I
42 0.32 A min I
43 0.31 A min I
44 0.33 A min I
45 0.33 A min I
46 0.31 A min I
47 0.3 A min I
48 0.33 A min I
49 0.34 A min I
50 0.3 A min I
51 0.32 A min I
52 0.33 A min I
53 0.32 A min I
54 0.3 A min I
55 0.32 A min I
56 0.28 A min I
57 0.3 A min I
58 0.25 A min I
59 0.26 A min I
60 0.26 A min I
61 0.3 A min I
62 0.32 A min I
63 0.3 A min I
64 0.29 A min I
65 0.3 A min I
66 0.32 B min I
67 0.31 B min I
68 0.3 B min I
69 0.27 B min I
70 0.24 B min I
71 0.27 B min I
72 0.27 B min I
73 0.25 B min I
74 0.28 B min I
75 0.27 B min I
76 0.28 B min I
77 0.23 B min I
78 0.27 B min I
79 0.27 B min I
80 0.28 B min I
81 0.3 B min I
82 0.3 B min I
83 0.33 B min I
84 0.3 B min I
85 0.29 B min I
86 0.31 B min I
87 0.3 B min I
88 0.33 B min I
89 0.35 B min I
90 0.35 B min I
91 0.36 B min I
92 0.34 B min I
93 0.35 B min I
94 0.35 B min I
95 0.33 B min I
96 0.3 B min I
97 0.33 B min I
98 0.3 B min I
99 0.33 B min I
100 0.34 B min I
101 0.3 B min I
102 0.33 B min I
103 0.3 B min I
104 0.32 B min I
105 0.34 B min I
106 0.4 B min I
107 0.29 B min I
108 0.31 B min I
109 0.32 B min I
110 0.34 B min I
111 0.33 B min I
112 0.31 B min I
113 0.29 B min I
114 0.3 B min I
115 0.33 B min I
116 0.33 B min I
117 0.34 B min I
118 0.31 B min I
119 0.34 B min I
120 0.35 B min I
121 0.29 B min I
122 0.28 B min I
123 0.26 B min I
124 0.27 B min I
125 0.26 B min I
126 0.32 B min I
127 0.33 B min I
128 0.33 B min I
129 0.32 B min I
130 0.34 B min I
131 0.46 A max I
132 0.39 A max I
133 0.44 A max I
134 0.48 A max I
135 0.5 A max I
136 0.43 A max I
137 0.44 A max I
138 0.45 A max I
139 0.44 A max I
140 0.41 A max I
141 0.44 A max I
142 0.47 A max I
143 0.42 A max I
144 0.45 A max I
145 0.46 A max I
146 0.4 A max I
147 0.39 A max I
148 0.42 A max I
149 0.42 A max I
150 0.43 A max I
151 0.4 A max I
152 0.37 A max I
153 0.42 A max I
154 0.39 A max I
155 0.41 A max I
156 0.4 A max I
157 0.43 A max I
158 0.43 A max I
159 0.42 A max I
160 0.45 A max I
161 0.42 A max I
162 0.42 A max I
163 0.41 A max I
164 0.42 A max I
165 0.45 A max I
166 0.41 A max I
167 0.38 A max I
168 0.43 A max I
169 0.41 A max I
170 0.39 A max I
171 0.37 A max I
172 0.41 A max I
173 0.39 A max I
174 0.44 A max I
175 0.4 A max I
176 0.41 A max I
177 0.42 A max I
178 0.39 A max I
179 0.43 A max I
180 0.4 A max I
181 0.43 A max I
182 0.46 A max I
183 0.44 A max I
184 0.43 A max I
185 0.39 A max I
186 0.4 A max I
187 0.4 A max I
188 0.43 A max I
189 0.42 A max I
190 0.43 A max I
191 0.4 A max I
192 0.45 A max I
193 0.43 A max I
194 0.42 A max I
195 0.43 A max I
196 0.45 B max I
197 0.46 B max I
198 0.42 B max I
199 0.49 B max I
200 0.5 B max I
201 0.42 B max I
202 0.43 B max I
203 0.45 B max I
204 0.43 B max I
205 0.46 B max I
206 0.53 B max I
207 0.52 B max I
208 0.46 B max I
209 0.52 B max I
210 0.48 B max I
211 0.49 B max I
212 0.47 B max I
213 0.46 B max I
214 0.49 B max I
215 0.5 B max I
216 0.46 B max I
217 0.45 B max I
218 0.45 B max I
219 0.46 B max I
220 0.49 B max I
221 0.48 B max I
222 0.49 B max I
223 0.49 B max I
224 0.48 B max I
225 0.49 B max I
226 0.48 B max I
227 0.47 B max I
228 0.49 B max I
229 0.49 B max I
230 0.47 B max I
231 0.46 B max I
232 0.44 B max I
233 0.45 B max I
234 0.44 B max I
235 0.42 B max I
236 0.39 B max I
237 0.44 B max I
238 0.44 B max I
239 0.43 B max I
240 0.42 B max I
241 0.41 B max I
242 0.42 B max I
243 0.43 B max I
244 0.39 B max I
245 0.44 B max I
246 0.42 B max I
247 0.41 B max I
248 0.41 B max I
249 0.44 B max I
250 0.43 B max I
251 0.46 B max I
252 0.46 B max I
253 0.52 B max I
254 0.48 B max I
255 0.49 B max I
256 0.48 B max I
257 0.5 B max I
258 0.49 B max I
259 0.47 B max I
260 0.5 B max I
261 0.43 A min E
262 0.45 A min E
263 0.43 A min E
264 0.42 A min E
265 0.4 A min E
266 0.41 A min E
267 0.43 A min E
268 0.41 A min E
269 0.44 A min E
270 0.4 A min E
271 0.43 A min E
272 0.42 A min E
273 0.4 A min E
274 0.42 A min E
275 0.4 A min E
276 0.39 A min E
277 0.41 A min E
278 0.4 A min E
279 0.41 A min E
280 0.39 A min E
281 0.39 A min E
282 0.37 A min E
283 0.37 A min E
284 0.36 A min E
285 0.37 A min E
286 0.39 A min E
287 0.37 A min E
288 0.39 A min E
289 0.39 A min E
290 0.36 A min E
291 0.4 A min E
292 0.38 A min E
293 0.4 A min E
294 0.37 A min E
295 0.36 A min E
296 0.37 A min E
297 0.39 A min E
298 0.39 A min E
299 0.38 A min E
300 0.36 A min E
301 0.4 A min E
302 0.39 A min E
303 0.39 A min E
304 0.37 A min E
305 0.36 A min E
306 0.38 A min E
307 0.39 A min E
308 0.37 A min E
309 0.39 A min E
310 0.41 A min E
311 0.36 A min E
312 0.39 A min E
313 0.37 A min E
314 0.37 A min E
315 0.38 A min E
316 0.39 A min E
317 0.4 A min E
318 0.43 A min E
319 0.4 A min E
320 0.39 A min E
321 0.37 A min E
322 0.36 A min E
323 0.38 A min E
324 0.36 A min E
325 0.35 A min E
326 0.32 B min E
327 0.33 B min E
328 0.34 B min E
329 0.4 B min E
330 0.32 B min E
331 0.34 B min E
332 0.34 B min E
333 0.35 B min E
334 0.34 B min E
335 0.31 B min E
336 0.32 B min E
337 0.35 B min E
338 0.34 B min E
339 0.4 B min E
340 0.36 B min E
341 0.38 B min E
342 0.39 B min E
343 0.39 B min E
344 0.37 B min E
345 0.38 B min E
346 0.38 B min E
347 0.35 B min E
348 0.39 B min E
349 0.36 B min E
350 0.37 B min E
351 0.36 B min E
352 0.39 B min E
353 0.38 B min E
354 0.37 B min E
355 0.36 B min E
356 0.38 B min E
357 0.36 B min E
358 0.35 B min E
359 0.37 B min E
360 0.38 B min E
361 0.3 B min E
362 0.32 B min E
363 0.32 B min E
364 0.34 B min E
365 0.29 B min E
366 0.38 B min E
367 0.36 B min E
368 0.38 B min E
369 0.35 B min E
370 0.36 B min E
371 0.36 B min E
372 0.34 B min E
373 0.34 B min E
374 0.33 B min E
375 0.29 B min E
376 0.35 B min E
377 0.37 B min E
378 0.33 B min E
379 0.36 B min E
380 0.34 B min E
381 0.33 B min E
382 0.33 B min E
383 0.3 B min E
384 0.32 B min E
385 0.33 B min E
386 0.31 B min E
387 0.32 B min E
388 0.32 B min E
389 0.33 B min E
390 0.33 B min E
391 0.48 A max E
392 0.51 A max E
393 0.49 A max E
394 0.5 A max E
395 0.53 A max E
396 0.5 A max E
397 0.47 A max E
398 0.46 A max E
399 0.5 A max E
400 0.48 A max E
401 0.47 A max E
402 0.5 A max E
403 0.5 A max E
404 0.51 A max E
405 0.46 A max E
406 0.47 A max E
407 0.47 A max E
408 0.49 A max E
409 0.5 A max E
410 0.47 A max E
411 0.45 A max E
412 0.48 A max E
413 0.44 A max E
414 0.46 A max E
415 0.48 A max E
416 0.47 A max E
417 0.45 A max E
418 0.48 A max E
419 0.46 A max E
420 0.48 A max E
421 0.47 A max E
422 0.45 A max E
423 0.47 A max E
424 0.46 A max E
425 0.49 A max E
426 0.45 A max E
427 0.49 A max E
428 0.46 A max E
429 0.49 A max E
430 0.47 A max E
431 0.48 A max E
432 0.51 A max E
433 0.52 A max E
434 0.48 A max E
435 0.48 A max E
436 0.45 A max E
437 0.51 A max E
438 0.46 A max E
439 0.46 A max E
440 0.53 A max E
441 0.46 A max E
442 0.51 A max E
443 0.48 A max E
444 0.47 A max E
445 0.45 A max E
446 0.47 A max E
447 0.46 A max E
448 0.52 A max E
449 0.5 A max E
450 0.52 A max E
451 0.48 A max E
452 0.49 A max E
453 0.48 A max E
454 0.46 A max E
455 0.47 A max E
456 0.49 B max E
457 0.45 B max E
458 0.48 B max E
459 0.48 B max E
460 0.51 B max E
461 0.46 B max E
462 0.45 B max E
463 0.51 B max E
464 0.44 B max E
465 0.5 B max E
466 0.51 B max E
467 0.52 B max E
468 0.53 B max E
469 0.47 B max E
470 0.48 B max E
471 0.53 B max E
472 0.55 B max E
473 0.53 B max E
474 0.56 B max E
475 0.55 B max E
476 0.53 B max E
477 0.53 B max E
478 0.49 B max E
479 0.49 B max E
480 0.47 B max E
481 0.51 B max E
482 0.55 B max E
483 0.51 B max E
484 0.54 B max E
485 0.55 B max E
486 0.5 B max E
487 0.48 B max E
488 0.52 B max E
489 0.5 B max E
490 0.47 B max E
491 0.46 B max E
492 0.52 B max E
493 0.51 B max E
494 0.49 B max E
495 0.48 B max E
496 0.46 B max E
497 0.51 B max E
498 0.48 B max E
499 0.5 B max E
500 0.52 B max E
501 0.46 B max E
502 0.49 B max E
503 0.54 B max E
504 0.51 B max E
505 0.48 B max E
506 0.49 B max E
507 0.48 B max E
508 0.5 B max E
509 0.49 B max E
510 0.48 B max E
511 0.49 B max E
512 0.54 B max E
513 0.49 B max E
514 0.5 B max E
515 0.5 B max E
516 0.51 B max E
517 0.51 B max E
518 0.5 B max E
519 0.48 B max E
520 0.49 B max E
DF <- data.frame(Nr_crt = 1:10,
AImin = round(rnorm(10),2),
BImin = round(rnorm(10),2),
AImax = round(rnorm(10),2),
BImax = round(rnorm(10),2),
AEmin = round(rnorm(10),2),
BEmin = round(rnorm(10),2),
AEmax = round(rnorm(10),2),
BEmax = round(rnorm(10),2))
DF
#The results should be ;See below
Nr _crt AImin BImin AImax BImax AEmin BEmin AEmax BEmax
1 0.31 0.32 0.46 0.45 0.43 0.32 0.48 0.49
2 0.34 0.31 0.39 0.46 0.45 0.33 0.51 0.45
3 0.37 0.3 0.44 0.42 0.43 0.34 0.49 0.48
4 0.3 0.27 0.48 0.49 0.42 0.4 0.5 0.48
5 0.28 0.24 0.5 0.5 0.4 0.32 0.53 0.51
6 0.31 0.27 0.43 0.42 0.41 0.34 0.5 0.46
7 0.34 0.27 0.44 0.43 0.43 0.34 0.47 0.45
8 0.35 0.25 0.45 0.45 0.41 0.35 0.46 0.51
9 0.32 0.28 0.44 0.43 0.44 0.34 0.5 0.44
10 0.35 0.27 0.41 0.46 0.4 0.31 0.48 0.5
11 0.27 0.28 0.44 0.53 0.43 0.32 0.47 0.51
12 0.29 0.23 0.47 0.52 0.42 0.35 0.5 0.52
13 0.31 0.27 0.42 0.46 0.4 0.34 0.5 0.53
14 0.28 0.27 0.45 0.52 0.42 0.4 0.51 0.47
15 0.3 0.28 0.46 0.48 0.4 0.36 0.46 0.48
16 0.29 0.3 0.4 0.49 0.39 0.38 0.47 0.53
17 0.29 0.3 0.39 0.47 0.41 0.39 0.47 0.55
18 0.3 0.33 0.42 0.46 0.4 0.39 0.49 0.53
19 0.32 0.3 0.42 0.49 0.41 0.37 0.5 0.56
20 0.29 0.29 0.43 0.5 0.39 0.38 0.47 0.55
21 0.33 0.31 0.4 0.46 0.39 0.38 0.45 0.53
22 0.3 0.3 0.37 0.45 0.37 0.35 0.48 0.53
23 0.31 0.33 0.42 0.45 0.37 0.39 0.44 0.49
24 0.32 0.35 0.39 0.46 0.36 0.36 0.46 0.49
25 0.33 0.35 0.41 0.49 0.37 0.37 0.48 0.47
26 0.31 0.36 0.4 0.48 0.39 0.36 0.47 0.51
27 0.29 0.34 0.43 0.49 0.37 0.39 0.45 0.55
28 0.31 0.35 0.43 0.49 0.39 0.38 0.48 0.51
29 0.32 0.35 0.42 0.48 0.39 0.37 0.46 0.54
30 0.28 0.33 0.45 0.49 0.36 0.36 0.48 0.55
31 0.32 0.3 0.42 0.48 0.4 0.38 0.47 0.5
32 0.33 0.33 0.42 0.47 0.38 0.36 0.45 0.48
33 0.33 0.3 0.41 0.49 0.4 0.35 0.47 0.52
34 0.31 0.33 0.42 0.49 0.37 0.37 0.46 0.5
35 0.32 0.34 0.45 0.47 0.36 0.38 0.49 0.47
36 0.31 0.3 0.41 0.46 0.37 0.3 0.45 0.46
37 0.3 0.33 0.38 0.44 0.39 0.32 0.49 0.52
38 0.3 0.3 0.43 0.45 0.39 0.32 0.46 0.51
39 0.32 0.32 0.41 0.44 0.38 0.34 0.49 0.49
40 0.32 0.34 0.39 0.42 0.36 0.29 0.47 0.48
41 0.32 0.4 0.37 0.39 0.4 0.38 0.48 0.46
42 0.32 0.29 0.41 0.44 0.39 0.36 0.51 0.51
43 0.31 0.31 0.39 0.44 0.39 0.38 0.52 0.48
44 0.33 0.32 0.44 0.43 0.37 0.35 0.48 0.5
45 0.33 0.34 0.4 0.42 0.36 0.36 0.48 0.52
46 0.31 0.33 0.41 0.41 0.38 0.36 0.45 0.46
47 0.3 0.31 0.42 0.42 0.39 0.34 0.51 0.49
48 0.33 0.29 0.39 0.43 0.37 0.34 0.46 0.54
49 0.34 0.3 0.43 0.39 0.39 0.33 0.46 0.51
50 0.3 0.33 0.4 0.44 0.41 0.29 0.53 0.48
51 0.32 0.33 0.43 0.42 0.36 0.35 0.46 0.49
52 0.33 0.34 0.46 0.41 0.39 0.37 0.51 0.48
53 0.32 0.31 0.44 0.41 0.37 0.33 0.48 0.5
54 0.3 0.34 0.43 0.44 0.37 0.36 0.47 0.49
55 0.32 0.35 0.39 0.43 0.38 0.34 0.45 0.48
56 0.28 0.29 0.4 0.46 0.39 0.33 0.47 0.49
57 0.3 0.28 0.4 0.46 0.4 0.33 0.46 0.54
58 0.25 0.26 0.43 0.52 0.43 0.3 0.52 0.49
59 0.26 0.27 0.42 0.48 0.4 0.32 0.5 0.5
60 0.26 0.26 0.43 0.49 0.39 0.33 0.52 0.5
61 0.3 0.32 0.4 0.48 0.37 0.31 0.48 0.51
62 0.32 0.33 0.45 0.5 0.36 0.32 0.49 0.51
63 0.3 0.33 0.43 0.49 0.38 0.32 0.48 0.5
64 0.29 0.32 0.42 0.47 0.36 0.33 0.46 0.48
65 0.3 0.34 0.43 0.5 0.35 0.33 0.47 0.49[/quote]Your complaint seems to e that I have not used the data that are in your pdf files.That was intentional. The point of my code was to show how to reshape your data and I considered the data used to be irrelevant.
I copied the data you last posted into a text file and used that as the data for the following code.
library(dplyr)
library(tidyr)
DF <- read.csv("~/R/Play/Dummy.csv", sep = " ")
head(DF)
#> Nr_crt Val CAP Tip Poz
#> 1 1 0.31 A min I
#> 2 2 0.34 A min I
#> 3 3 0.37 A min I
#> 4 4 0.30 A min I
#> 5 5 0.28 A min I
#> 6 6 0.31 A min I
DFwide <- DF |> mutate(name = paste0(CAP, Poz, Tip)) |>
select(Val, name) |>
mutate(Nr_crt = rep(1:(nrow(DF)/8),8)) |>
pivot_wider(names_from = name, values_from = "Val")
DFwide
#> # A tibble: 65 x 9
#> Nr_crt AImin BImin AImax BImax AEmin BEmin AEmax BEmax
#> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0.31 0.32 0.46 0.45 0.43 0.32 0.48 0.49
#> 2 2 0.34 0.31 0.39 0.46 0.45 0.33 0.51 0.45
#> 3 3 0.37 0.3 0.44 0.42 0.43 0.34 0.49 0.48
#> 4 4 0.3 0.27 0.48 0.49 0.42 0.4 0.5 0.48
#> 5 5 0.28 0.24 0.5 0.5 0.4 0.32 0.53 0.51
#> 6 6 0.31 0.27 0.43 0.42 0.41 0.34 0.5 0.46
#> 7 7 0.34 0.27 0.44 0.43 0.43 0.34 0.47 0.45
#> 8 8 0.35 0.25 0.45 0.45 0.41 0.35 0.46 0.51
#> 9 9 0.32 0.28 0.44 0.43 0.44 0.34 0.5 0.44
#> 10 10 0.35 0.27 0.41 0.46 0.4 0.31 0.48 0.5
#> # ... with 55 more rows
Created on 2022-07-18 by the reprex package (v2.0.1)
I was wondering FJCC how could we count unique combinations of CAP, Tip, Poz ? It obviously gives 65 but how to do it with code and sort of a frequency table ? My apologise about a bit off topic.
@Andrzej - I don't understand your question. There are eight combinations of CAP, Tip and Poz, two values for each variable. CAP is A or B, Tip is min or max and Poz is I or E. Each of those combinations gets measured 65 times. In the wide data format, that 65 is obvious from the number of rows. In the long format, you have to know there are eight combinations to divide the 520 rows by 8 to get 65. You can get the eight combinations with
unique(DF[,c("CAP","Tip","Poz")])
if DF is the long form of the data.
which gives:
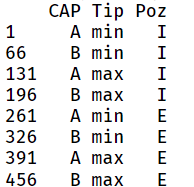
My desired output would be like this, please:
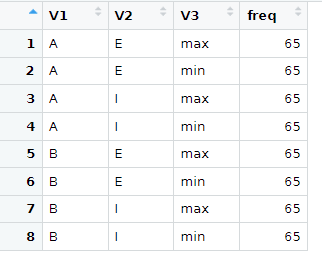
That would be this code, if DF is the long format.
library(dplyr)
> DF |> group_by(CAP,Tip,Poz) |> count()
# A tibble: 8 x 4
# Groups: CAP, Tip, Poz [8]
CAP Tip Poz n
<chr> <chr> <chr> <int>
1 A max E 65
2 A max I 65
3 A min E 65
4 A min I 65
5 B max E 65
6 B max I 65
7 B min E 65
8 B min I 65
Thank you very much indeed.
DFwide <- DF |> mutate(name = paste0(CAP, Poz, Tip)) |>
select(Val, name) |>
mutate(Nr_crt = rep(1:(nrow(DF)/8),8)) |>
pivot_wider(names_from = name, values_from = "Val")
DFwide
see error:
Error in pivot_wider(names_from = name, values_from = "Val") :
object 'name' not found
try putting name in speech marks
pivot_wider(names_from = "name", values_from = "Val")
if that doesn't work, or you have more issues/questions, please provide a reprex; This thread is now quite long and seems fragmented, it would be good to have a single post with example data and code that reproduces a problem in order to address it succinctly.
I gave up
Thnaks a lot for your time .
Do not work
This topic was automatically closed 21 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.