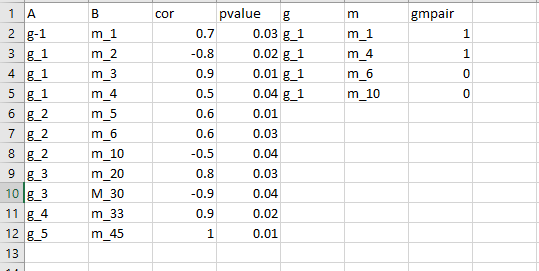
I am not quite sure if this something you want. But, you can see I have given two different types of solutions, first exactly like what you showed. I just put two datasets together side by side not considering anything and add an indicator variable if xx data set A, B matches to g m of another dataset. I think there were 4 of them that matched.
I also tried the merge approach. You see if there is no NA in a row that means the pair are present in two datasets otherwise they are in either of one xx or target. But they are not put together.
#####
library(tidyverse)
set.seed(42)
B <- matrix(rnorm(1000), nrow = 100)
rownames(B) <- paste0("m_", ncol = 1:100)
colnames(B) <- paste0("S_", ncol = 1:10)
set.seed(145)
A <- matrix(rnorm(1000), nrow = 100)
rownames(A) <- paste0("g_", ncol = 1:100)
colnames(A) <- paste0("S_", ncol = 1:10)
targetlist <- data.frame(g = sample(rownames(A),50), m = sample(rownames(B),50))
library(dplyr)
sig_correlation2Dfs<-function(A,B){
n <- t(!is.na(A)) %*% (!is.na(B))
r <- cor(A, B, use = "pairwise.complete.obs")
cor2pvalue = function(r, n) {
t <- (r*sqrt(n-2))/sqrt(1-r^2) ## t-distribution
p <- 2*(1 - pt(abs(t),(n-2))) ## pvalue
out <- list(r, n, t, p)
names(out) <- c("r", "n", "t", "p")
return(out)
}
# Get a list with matrices of correlation, pvalues
result = cor2pvalue(r,n)
rcoeffMatrix<-result$r
pvalueMatrix<-result$p
rows<-rownames(rcoeffMatrix)
cols<-colnames(pvalueMatrix)
df <- data.frame(A=character(),
B=character(),
cor=double(),
Pvalue=double(),
stringsAsFactors=FALSE)
for(i in rows){
for(j in cols){
if (pvalueMatrix[i,j] > 0.05){
next
}else{
cor<-rcoeffMatrix[i,j]
Pvalue<-pvalueMatrix[i,j]
df<- df %>% add_row(A = i, B = j, cor = cor, Pvalue = Pvalue)
}
}
}
df
}
xx <- sig_correlation2Dfs(t(A), t(B))
targetlist$g<- as.character(targetlist$g)
targetlist$m<- as.character(targetlist$m)
i1 <- match(paste(xx$A, xx$B), paste(targetlist$g, targetlist$m), nomatch = 0)
i1[i1>1] = 1
library(qpcR)
#> Loading required package: MASS
#>
#> Attaching package: 'MASS'
#> The following object is masked from 'package:dplyr':
#>
#> select
#> Loading required package: minpack.lm
#> Loading required package: rgl
#> Loading required package: robustbase
#> Loading required package: Matrix
#>
#> Attaching package: 'Matrix'
#> The following objects are masked from 'package:tidyr':
#>
#> expand, pack, unpack
merge<- qpcR:::cbind.na(xx, targetlist)
merge$gmpair<- i1
merge [1:15,]
#> A B cor Pvalue g m gmpair
#> 1 g_1 m_1 0.7014863 0.023777696 g_7 m_54 0
#> 2 g_1 m_11 0.6542419 0.040136845 g_34 m_42 0
#> 3 g_1 m_30 -0.6608990 0.037473787 g_28 m_6 0
#> 4 g_1 m_57 0.8268228 0.003174772 g_16 m_1 0
#> 5 g_1 m_84 -0.7585984 0.010971358 g_77 m_37 0
#> 6 g_2 m_35 0.7309656 0.016317876 g_66 m_9 0
#> 7 g_2 m_54 0.6626535 0.036792338 g_44 m_64 0
#> 8 g_2 m_86 -0.8376313 0.002487474 g_72 m_28 0
#> 9 g_2 m_92 -0.6982939 0.024704891 g_12 m_71 0
#> 10 g_3 m_56 -0.7484422 0.012765535 g_32 m_15 0
#> 11 g_3 m_62 0.6412703 0.045685764 g_3 m_3 0
#> 12 g_3 m_63 0.6505131 0.041682715 g_81 m_94 0
#> 13 g_3 m_68 0.7852333 0.007117049 g_18 m_32 0
#> 14 g_3 m_75 -0.7357742 0.015279416 g_55 m_29 0
#> 15 g_3 m_88 -0.6469525 0.043195695 g_91 m_65 0
# Another method
xx$ID1<- as.character(xx$A)
xx$ID2<- as.character(xx$B)
targetlist$ID1<- as.character(targetlist$g)
targetlist$ID2<- as.character(targetlist$m)
merge.file<- merge(xx, targetlist, by= c("ID1", "ID2"), all = TRUE )
merge.file$gmpair<- ifelse(complete.cases(merge.file)==1, 1,0)
merge.file<- merge.file %>% dplyr::select(-c(ID1, ID2))
merge.file[1:30,]
#> A B cor Pvalue g m gmpair
#> 1 g_1 m_1 0.7014863 0.0237776960 <NA> <NA> 0
#> 2 g_1 m_11 0.6542419 0.0401368454 <NA> <NA> 0
#> 3 g_1 m_30 -0.6608990 0.0374737872 <NA> <NA> 0
#> 4 g_1 m_57 0.8268228 0.0031747724 <NA> <NA> 0
#> 5 g_1 m_84 -0.7585984 0.0109713582 <NA> <NA> 0
#> 6 g_10 m_13 -0.9057432 0.0003077797 <NA> <NA> 0
#> 7 g_10 m_14 -0.8999465 0.0003879589 <NA> <NA> 0
#> 8 g_10 m_32 -0.6547675 0.0399220998 <NA> <NA> 0
#> 9 g_10 m_40 0.6407355 0.0459249438 <NA> <NA> 0
#> 10 g_10 m_47 0.7228194 0.0181874913 <NA> <NA> 0
#> 11 <NA> <NA> NA NA g_10 m_79 0
#> 12 g_10 m_83 -0.7845922 0.0071963977 <NA> <NA> 0
#> 13 g_100 m_32 0.6345359 0.0487590474 <NA> <NA> 0
#> 14 g_100 m_46 -0.6374117 0.0474302990 <NA> <NA> 0
#> 15 g_100 m_67 0.6602530 0.0377268136 <NA> <NA> 0
#> 16 g_100 m_73 0.8017408 0.0052803843 <NA> <NA> 0
#> 17 g_100 m_86 0.6658360 0.0355776974 <NA> <NA> 0
#> 18 g_11 m_12 -0.6338640 0.0490730266 <NA> <NA> 0
#> 19 g_11 m_45 0.7271768 0.0171699107 <NA> <NA> 0
#> 20 g_11 m_67 0.6687339 0.0344955594 <NA> <NA> 0
#> 21 g_11 m_8 0.7732543 0.0087056588 <NA> <NA> 0
#> 22 g_11 m_96 0.7467191 0.0130890753 <NA> <NA> 0
#> 23 g_11 m_97 0.7724212 0.0088246927 <NA> <NA> 0
#> 24 g_12 m_1 -0.6434832 0.0447049110 <NA> <NA> 0
#> 25 g_12 m_2 0.6809475 0.0301796889 <NA> <NA> 0
#> 26 g_12 m_22 0.7979446 0.0056686682 <NA> <NA> 0
#> 27 g_12 m_43 -0.6561107 0.0393767892 <NA> <NA> 0
#> 28 g_12 m_7 -0.6621928 0.0369704595 <NA> <NA> 0
#> 29 <NA> <NA> NA NA g_12 m_71 0
#> 30 g_12 m_9 -0.7006940 0.0240055301 <NA> <NA> 0