Hi, this is my first time posting on here so please excuse any mistakes I might make.
I am currently writing my final code for my dissertation and I wanted to make my maps look neat together, So I need to make their legends universal.
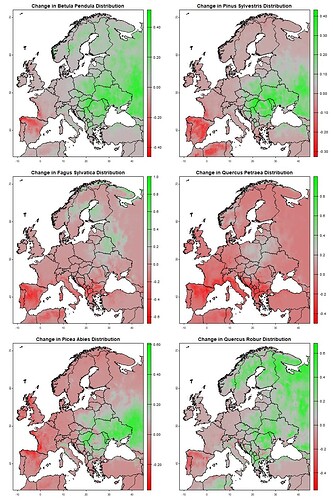
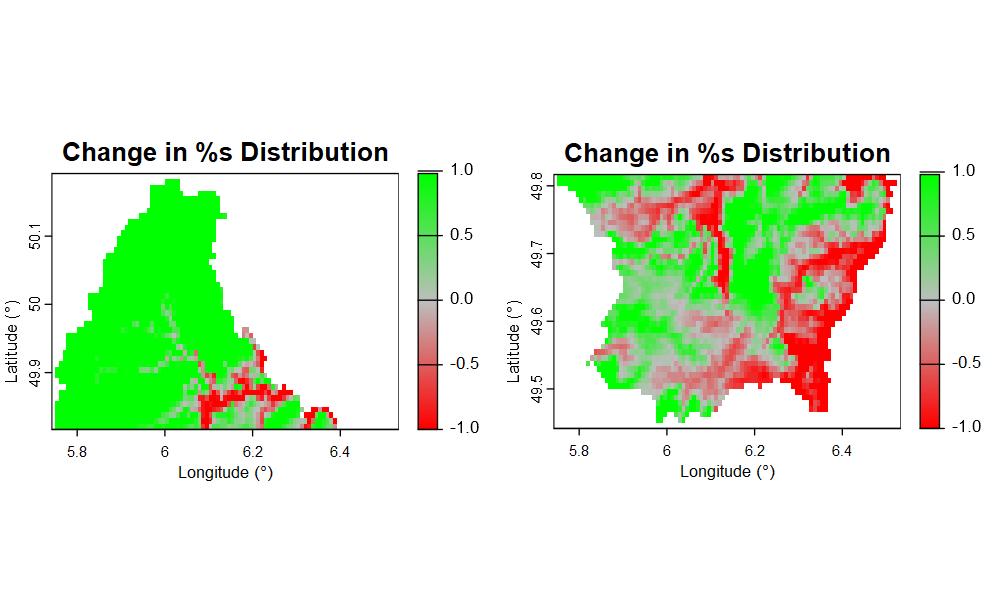
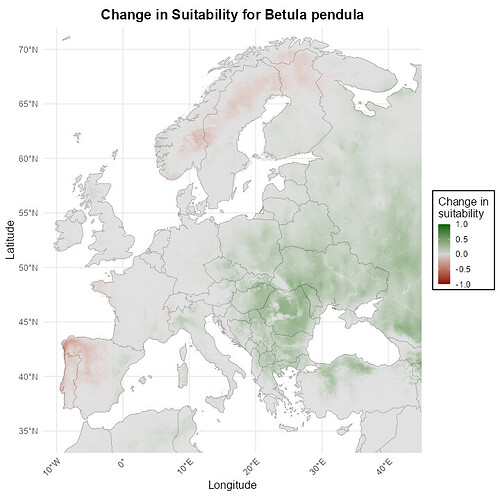
As you can see in the image, none of the legends align up with each other. I aim to have grey represent 0, the greenest 1 and the redest -1, but it hasnt worked well for my maps.
And if it would be possible, how could I add a legend title next to the gradient to desribe what it is?
My code for the maps:
(I do have more code for the species downloading, cleaning etc)
# Calculate the difference in predicted suitability
suitability_change <- forecast_presence - glm_predict
# Mask values outside the range [-1, 1]
suitability_change_limited <- mask(suitability_change, suitability_change >= -1 & suitability_change <= 1)
# Number of breaks
n_breaks <- 100
# Assuming your breaks are correctly set for the range [-1, 1]
breaks <- seq(-1, 1, length.out = n_breaks + 1) # Recalculate breaks for the limited range
classified <- classify(suitability_change_limited, rcl = cbind(breaks, 1:length(breaks)))
# Generate a color gradient for the limited range
colors <- colorRampPalette(c("red", "grey", "green"))(n_breaks)
# Proceed with plotting
base_map_filename <- sprintf("output/Change in %s Distribution.png", species_common_name)
png(base_map_filename, width = 720, height = 720)
plot(classified,
axes=TRUE,
col=colors,
xlab = "Longitude (°)",
ylab = "Latitude (°)",
main= sprintf("Change in %s Distribution", species_common_name))
plot(my_map, add = TRUE, border = "grey5")
dev.off()