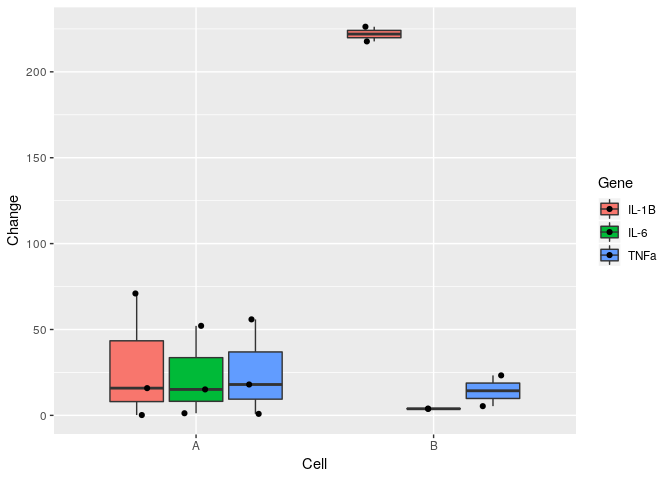
Hi there, in my dataset I have two cell types that had three genes tested on each.
I want to obtain a grouped barplot/boxplot, importantly, I want to be able to visualise individual data points on the bar/box.
cell <- c(1,1,1,1,1,1,1,1,1,2,2,2,2,2,2)
Gene <- c("IL-6","IL-6","IL-6","IL-1B","IL-1B","IL-1B","TNFa","TNFa","TNFa","IL-6","IL-6","IL-1B","IL-1B","TNFa","TNFa")
Change <- c(52.1204352,15.0980129,1.2306710,70.9649383,15.8489764,0.1797975,55.8863164,17.9731222,0.8732369,3.7435065,3.9501774,217.7251295,226.2724972,23.2779805,5.3588277)
Where I got so far:
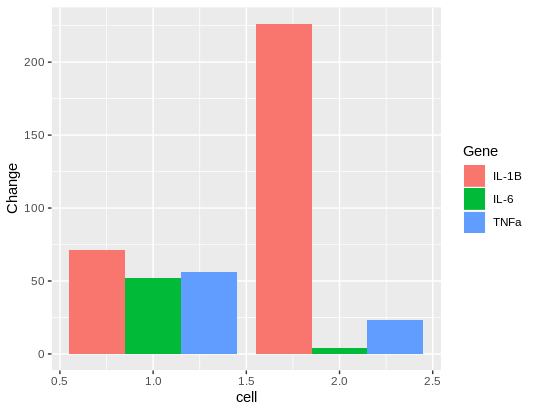
ggplot(df, aes(y = Change,
x = cell,
group = Gene)) +
geom_bar(stat = "identity", position = "dodge", aes(fill = Gene))
so I got this
Could someone guide me in plotting the individual data points on top?
And, ideally, I would have Cell A and Cell B on x axis instead of numerical values, but that I can tweak later on Photoshop if I get the individual data points to come through.
Thank you so much!