I've been working with ggscatmat from the GGplot library to generate plots of a correlation matrix. I am running into two problems/questions:
-
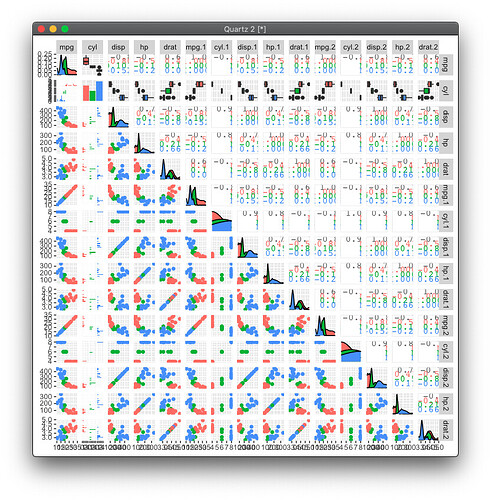
When I increase the column number in the plots to go beyond 9, the formatting of the plots gets messed up. Is there a hard limit to the pairs of columns I can compare?
-
I'm getting a warning that
Factor variables are omitted in plot. However, all columns in the dataset are numeric, so I am puzzled why this warning appears.
Any thoughts?
#// load GGally library
library(GGally)
#> Loading required package: ggplot2
#> Registered S3 method overwritten by 'GGally':
#> method from
#> +.gg ggplot2
#// generate datafram
data <- data.frame(rep(mtcars[1:5], 3))
#// ggscatmat with increasing column numbers
ggscatmat(data[1:5], columns = 1:5, color = "cyl", alpha = 0.8)
#> Warning in ggscatmat(data[1:5], columns = 1:5, color = "cyl", alpha = 0.8):
#> Factor variables are omitted in plot
[new user only allowed 1 plot per post]
ggscatmat(data[1:10], columns = 1:10, color = "cyl", alpha = 0.8)
#> Warning in cor(xvalue, yvalue, use = "pairwise.complete.obs", method =
#> corMethod): the standard deviation is zero
[...]
#> Warning in ggscatmat(data[1:10], columns = 1:10, color = "cyl", alpha = 0.8):
#> Factor variables are omitted in plot
[new user only allowed 1 plot per post]
ggscatmat(data[1:15], columns = 1:15, color = "cyl", alpha = 0.8)
#> Warning in cor(xvalue, yvalue, use = "pairwise.complete.obs", method =
#> corMethod): the standard deviation is zero
[...]
#> Warning in ggscatmat(data[1:15], columns = 1:15, color = "cyl", alpha = 0.8):
#> Factor variables are omitted in plot
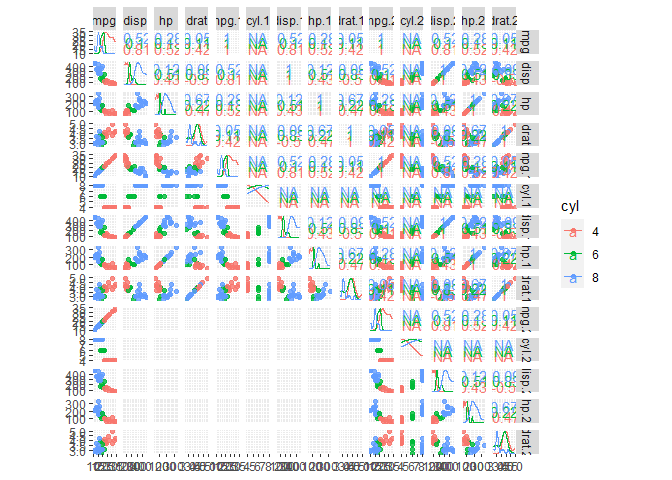
Created on 2021-01-29 by the reprex package (v0.3.0)