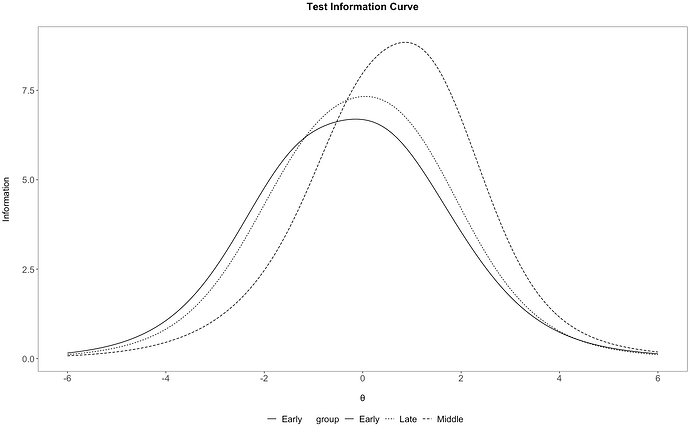
I think I figured out the mystery of the label-changing behavior: The function multipleGroups() will determine a group ordering implicitly if the group vector is not a factor, and will use the factor level ordering otherwise. In your original post, @baglolese, you supplied the group labels as a character vector, so multipleGroups() implicitly ordered them alphabetically. Later, the factor structure is not
automatically incorporated into the table tI in @raytong's code, so must be restored before plotting.
Below is an annotated and modified version of @raytong's code that incorporates these observations; it's written from a didactic point of view since I wasn't sure how familiar you are with the tidyverse .
library(haven)
library(tidyverse)
library(mirt)
newdat <- read_sav("IRTsampledata.sav")
newdata %>% head()
# create 'group' factor for use with 'multipleGroup()'
## extract indices of age range labels
age_indices <-
newdat %>%
pull(age)
age_indices %>% head()
## use indices to create 'group' vector of new age range labels
groups <- c("Early", 'Middle', "Late")
group <- groups[age_indices]
## convert to factor: necessary for use with 'multipleGroup()' to avoid
## order of groups being implicitly determined
group <- factor(group, levels = groups)
# alternative approach to creating 'group' factor in 'newdat' itself
newdat <-
newdat %>%
# extract original age range labels and indices
mutate(age_range = as_factor(age, levels = 'labels')) %>%
mutate(age_index = as_factor(age, levels = 'values'))
newdat %>% head()
## create column with new age range labels
newdat <-
newdat %>%
mutate(age_label =
case_when(
age_index == 1 ~ "Early",
age_index == 2 ~ "Middle",
age_index == 3 ~ "Late"
)
)
newdat %>% head()
## drop age_index and age_range columns, and reorder columns
newdat <-
newdat %>%
select(age_label, Item_1:Item_20)
newdat %>% head()
## extract age_label column data and compare with 'group'
group.dup <- newdat %>% pull(age_label)
all(group == group.dup)
# create multigroup object
mg <-
multipleGroup(
newdat %>% select(-age_label),
mirt.model('F1 = 1-20'),
group
)
## check ordering used for 'group' values
mg@Data$groupNames
# create table of results
Theta <- matrix(seq(-6,6,.01))
tI <- map_dfr(1:3, ~{
ti <- testinfo(mg, Theta, group = .x)
tibble(
information = ti,
xAxis = Theta,
se = 1/(sqrt(ti)),
group = groups[.x]
)
})
tI %>% head()
## Note: 'group' column is no longer a factor, so ggplot will use
## alphabetical order implicitly in legend
# set theme for IRT plot separately
irt_theme <-
theme_bw() +
theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_blank(),
axis.text=element_text(size=14),
axis.title=element_text(size=14),
legend.text=element_text(size=14),
legend.title=element_text(size=14),
plot.title = element_text(hjust = 0.5, size=16, face="bold"),
legend.position="bottom"
)
# create plot with 'tI' without converting 'group' to factor
tI %>%
ggplot(aes(x=xAxis, y=information)) +
# add lines, one per group value, distinguished by line type
geom_line(aes(linetype = group)) +
irt_theme +
# set labels and axis appearance
labs(x="\nθ", y = "Information\n") +
scale_x_continuous(breaks=c(-6,-4,-2,0,2,4,6)) +
ggtitle("Test Information Curve\n")
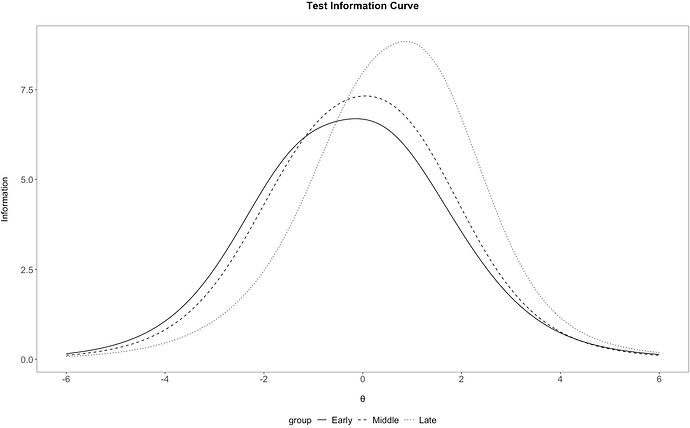
# create plot with 'tI', but converting 'group' to factor
## perform conversion in 'tI' itself
tI %>%
mutate(group = factor(group, levels = groups)) %>%
ggplot(aes(x=xAxis, y=information)) +
geom_line(aes(linetype = group)) +
irt_theme +
labs(x="\nθ", y = "Information\n") +
scale_x_continuous(breaks=c(-6,-4,-2,0,2,4,6)) +
ggtitle("Test Information Curve\n")
## alternatively, perform conversion when building 'tI'
tI <- map_dfr(1:3, ~{
ti <- testinfo(mg, Theta, group = .x)
tibble(
information = ti,
xAxis = Theta,
se = 1/(sqrt(ti)),
group = groups[.x] %>% factor(levels = groups)
)
})
tI %>% head()
tI %>%
ggplot(aes(x=xAxis, y=information)) +
geom_line(aes(linetype = group)) +
irt_theme +
labs(x="\nθ", y = "Information\n") +
scale_x_continuous(breaks=c(-6,-4,-2,0,2,4,6)) +
ggtitle("Test Information Curve\n")
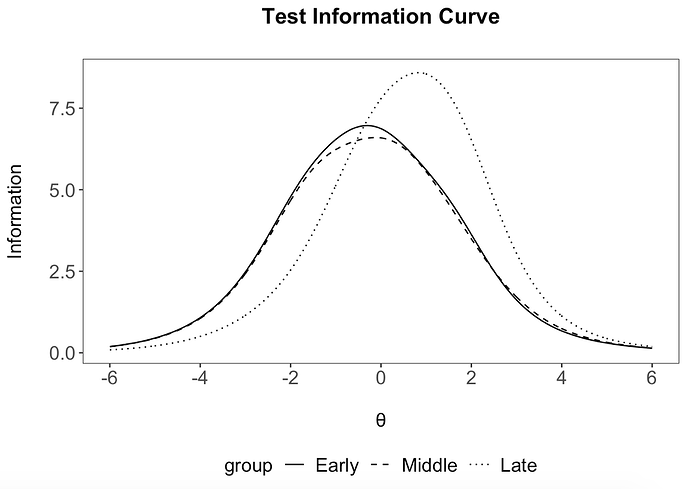
# exclude 'Late' group
tI %>%
mutate(group = factor(group, levels = groups)) %>%
filter(group != 'Late') %>%
ggplot(aes(x=xAxis, y=information)) +
geom_line(aes(linetype = group)) +
irt_theme +
labs(x="\nθ", y = "Information\n") +
scale_x_continuous(breaks=c(-6,-4,-2,0,2,4,6)) +
ggtitle("Test Information Curve\n")
![]()