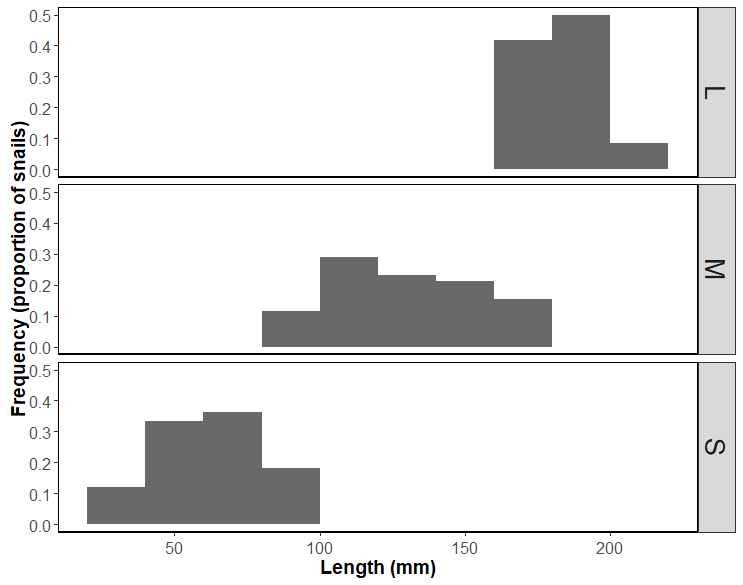
I am using code from a previous project and am receiving the below warning messages and the output is not showing different coloured bars or legend. Can someone please advise edits to code?
warning message:
Warning messages:
1: The following aesthetics were dropped during statistical transformation: fill.
![]() This can happen when ggplot fails to infer the correct grouping structure in the data.
This can happen when ggplot fails to infer the correct grouping structure in the data.
![]() Did you forget to specify a
Did you forget to specify a group aesthetic or to convert a numerical variable into a factor?
2: The following aesthetics were dropped during statistical transformation: fill.
![]() This can happen when ggplot fails to infer the correct grouping structure in the data.
This can happen when ggplot fails to infer the correct grouping structure in the data.
![]() Did you forget to specify a
Did you forget to specify a group aesthetic or to convert a numerical variable into a factor?
3: The following aesthetics were dropped during statistical transformation: fill.
![]() This can happen when ggplot fails to infer the correct grouping structure in the data.
This can happen when ggplot fails to infer the correct grouping structure in the data.
![]() Did you forget to specify a
Did you forget to specify a group aesthetic or to convert a numerical variable into a factor?
code:
ggplot(data=snail.data.qn, aes(length, y=20*after_stat(density), fill=year.factor))+
geom_histogram(position='dodge', binwidth=20, boundary=0, alpha=0.9)+
facet_grid(size~.)+
xlab("Length (mm)") +
ylab("Frequency (proportion of snails)") +
theme_bw() +
theme(panel.grid.minor=element_blank(), panel.grid.major=element_blank()) +
theme(panel.border = element_rect(linewidth=1, color = "black"))+
theme(legend.background = element_rect())+
theme(legend.justification=c(1,1), legend.position.inside=c(0.10,0.99)) +
theme(legend.title = element_text(size=14, face="bold")) +
theme(legend.text = element_text(size = 12)) +
theme(strip.text.x = element_text(size=20),
strip.text.y = element_text(size=20))+
theme(axis.title.x = element_text(face="bold", size=14),
axis.text.x = element_text(vjust=0.5, size=12))+
theme(axis.title.y = element_text(angle=90, vjust=0.45, face="bold", size=14),
axis.text.y = element_text(size=12))
Output: