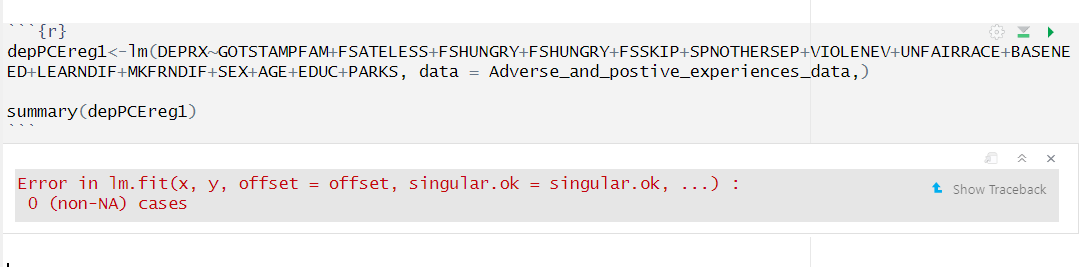
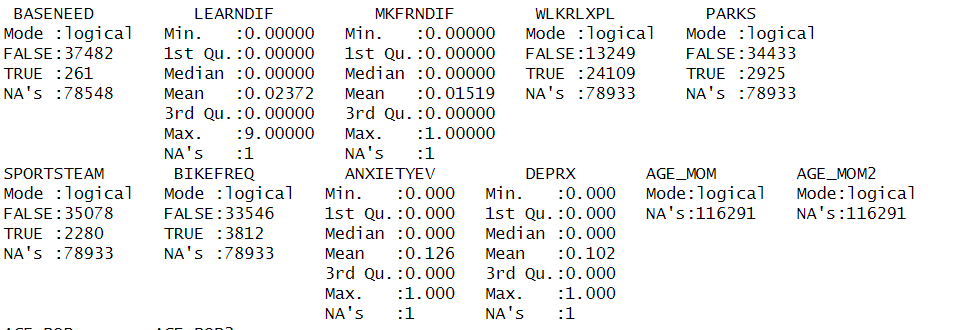
Hi everyone, This is an error that I keep getting because of the parks variable. There are four other variables similar to this one in my dataset and they all return the same error. My data is from an excel file and I know that normally this error would be returned if there was no data in the column, but there is at least 40,000 0 or 1 variables out of 100,000. I am not sure if I need to add a library or something, I had this issue earlier and just shaved down the dataset and that fixed it for the other ones. I can't really do that again because then I will lose other important variables altogether
Are you sure all the variables have come in from Excel as numeric?
Hi, I just checked and it looks like they did not. Is there a way to fix that too and will that resolve the issue? I used the import from excel function in r , I think it was read xl
You have a lot of missing data. You should check if there is any row where all of the variables used in the regression are not NA. Make a new data frame selecting only the columns used in the regression and then use the na.omit() function to find the rows where there are no NA values. For example, if you were just using the three columns "DEPRX","GOTSTAMPFAM","FSATELESS"
NewDF <- Adverse_and_postive_experience_data[, c("DEPRX","GOTSTAMPFAM","FSATELESS")
NewDF <- na.omit(NewDF)
nrow(NewDF)
The nrow() function will tell you how many rows have complete data.
What @FJCC says is good advice, but it does depend on whether those NA observations are really missing. Find a few of the NA observations and then go back and look carefully at the Excel spreadsheet to see if the value there should be legit but is somehow getting read in wrong.
# create a data frame and sprinkle NAs
dirty <- as.matrix(mtcars)
size <- dim(dirty)[1]*dim(dirty)[2]
ind <- sample(1:size, floor(0.1*size))
dirty[ind] <- NA
dirty <- as.data.frame(dirty)
colnames(dirty) <- paste0("V",1:11)
rownames(dirty) <- NULL
dirty
#> V1 V2 V3 V4 V5 V6 V7 V8 V9 V10 V11
#> 1 21.0 6 160.0 NA 3.90 2.620 16.46 0 NA 4 4
#> 2 21.0 NA 160.0 110 3.90 NA 17.02 0 1 4 4
#> 3 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
#> 4 21.4 NA 258.0 110 3.08 3.215 19.44 1 0 3 1
#> 5 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2
#> 6 18.1 6 225.0 105 2.76 3.460 20.22 1 0 NA 1
#> 7 14.3 NA 360.0 NA 3.21 3.570 NA 0 0 3 4
#> 8 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
#> 9 NA 4 140.8 95 NA 3.150 22.90 1 0 4 2
#> 10 19.2 6 167.6 123 3.92 3.440 18.30 1 0 NA 4
#> 11 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4
#> 12 16.4 8 275.8 180 NA NA NA 0 0 3 3
#> 13 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3
#> 14 15.2 8 275.8 180 NA 3.780 18.00 0 0 3 3
#> 15 10.4 8 NA 205 2.93 5.250 17.98 0 0 NA NA
#> 16 10.4 8 460.0 NA 3.00 5.424 17.82 0 0 3 4
#> 17 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4
#> 18 32.4 4 78.7 NA 4.08 2.200 19.47 1 1 4 1
#> 19 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
#> 20 33.9 4 71.1 65 4.22 1.835 19.90 NA 1 4 1
#> 21 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
#> 22 15.5 8 318.0 150 2.76 3.520 16.87 NA 0 3 2
#> 23 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2
#> 24 NA 8 350.0 245 3.73 3.840 15.41 0 0 3 4
#> 25 19.2 8 NA 175 3.08 3.845 NA 0 0 3 2
#> 26 27.3 4 NA 66 4.08 NA 18.90 1 1 4 1
#> 27 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
#> 28 30.4 4 NA 113 3.77 1.513 16.90 1 1 5 2
#> 29 15.8 8 351.0 264 4.22 3.170 NA 0 1 5 4
#> 30 19.7 NA 145.0 175 3.62 2.770 15.50 0 1 5 6
#> 31 NA 8 301.0 335 3.54 NA 14.60 0 1 5 NA
#> 32 21.4 NA 121.0 109 4.11 2.780 18.60 1 1 4 2
# select only rows without one or more NA
(clean <- dirty[complete.cases(dirty),])
#> V1 V2 V3 V4 V5 V6 V7 V8 V9 V10 V11
#> 3 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
#> 5 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2
#> 8 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
#> 11 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4
#> 13 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3
#> 17 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4
#> 19 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
#> 21 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
#> 23 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2
#> 27 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
Created on 2023-04-25 with reprex v2.0.2
Hi FJCC, Thank you! I tried running that code, but it returned an error saying this:
Error: unexpected symbol in:
"NewDF <- Adverse_and_postive_experience_data[, c("SPORTSTEAM","WLKRLXPL","PARKS")
NewDF"
Typo? "Experiences" needs an "s" at the end?
Needs ]. The “N” at the beginning of the last line is the unexpected symbol. The error is almost always due to missing delimiters.
There is also a missing "s" in the data frame name. Not one of my best efforts!
it's all good! thank you so much!
This topic was automatically closed 42 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.