Hello! I am trying to create a map with covid data as attribute.
I downloaded the covid data from covid-19-data/public/data at master · owid/covid-19-data · GitHub, and I joined this data frame to the world sf object found in the spData library (an sf object containing a geometry column corresponding to each country and other data such as population, continent, country name etc.).
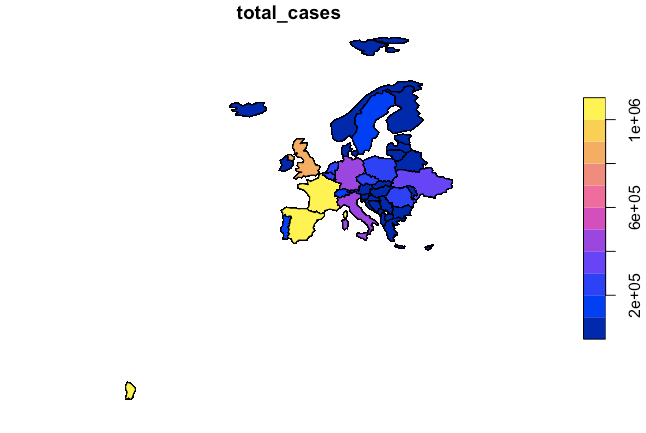
When I subset by Europe, I get in my map also some distant island (part of France), which I would like to remove (see below code and resulting plot). Does someone knows how I can do that? .. any suggestion would help!
thanks !
# Load libraries
library(sf)
library(raster)
library(spData)
install.packages("countrycode")
library(countrycode)
# Upload file
covid_dataset <- read.csv("~/Downloads/owid-covid-data (1).csv", header=TRUE)
str(covid_dataset)
head(covid_dataset)
# Change date format
covid_dataset$date <- as.Date(covid_dataset$date)
#To avoid confusion with country names, create a new variable with the same iso code
# Name the variable the same way for simplicity for joining
world_iso <- world %>%
mutate(iso_code = countryname(world$name_long, 'iso3c'))
world_iso %>% count(continent)
#Eliminate Antarctica
world_iso <- world_iso %>%
filter(continent != "Antarctica")
# Create subset with data feom March on
covid_dataset_from_march <- covid_dataset %>%
filter(date >= "2020-03-01")
# Merge world and covid_dataset
world_covid_data = left_join(world_iso, covid_dataset_from_march, by = "iso_code")
setdiff(world_iso$iso_code, covid_dataset_from_march$iso_code)
plot(world_covid_data['total_cases'])
# Remove some useless variables
world_covid_data <- world_covid_data %>%
dplyr::select(-continent.x, -location)
# Create subset with Europe (and exclude Russia)
europe_covid_data <- world_covid_data_2 %>%
filter(continent.y == "Europe")
europe_covid_data = filter(europe_covid_data, name_long != "Russian Federation")
plot(europe_covid_data['total_cases'])