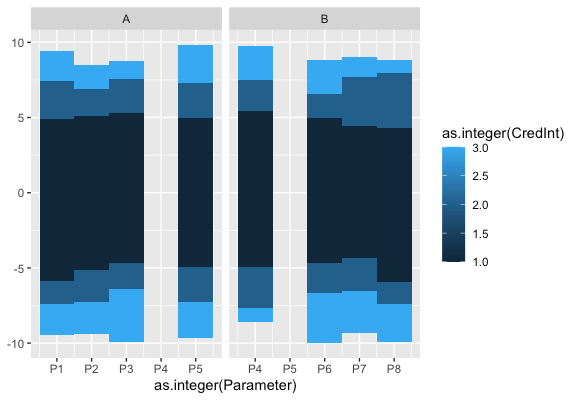
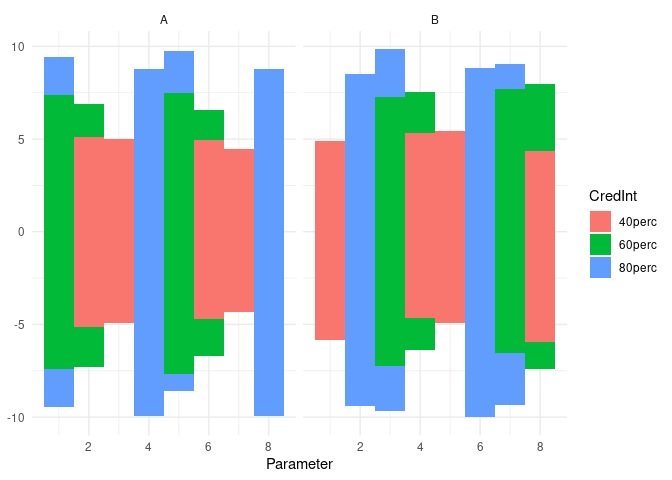
Edit: Added copy of plot generated
Hey,
Big fan of RStudio and ggplot, apologies for the clunky code below but hopefully it's reproducible.
I'm trying to plot credible intervals for a set of parameters and geom_rect seems to be the neatest way to plot them but means I have to treat the factors as integers but as soon as the factors are out of order things become messy.
Essentially, I want to be able to treat the x axis as a factor without doing the messy integer transformation back and forth.
An option to treat the x variable like geom_tile and the y variable like geom_rect seems to be ideal but I wondered if there's any alternative solution that's currently available.
library(ggplot2)
set.seed(1859)
dat <- data.frame("CredInt" = factor(rep(c("80perc", "60perc", "40perc"), 8)),
"Lower_CI" = c(runif(1, -10, -8), runif(1, -8, -6), runif(1, -6, -4),
runif(1, -10, -8), runif(1, -8, -6), runif(1, -6, -4),
runif(1, -10, -8), runif(1, -8, -6), runif(1, -6, -4),
runif(1, -10, -8), runif(1, -8, -6), runif(1, -6, -4),
runif(1, -10, -8), runif(1, -8, -6), runif(1, -6, -4),
runif(1, -10, -8), runif(1, -8, -6), runif(1, -6, -4),
runif(1, -10, -8), runif(1, -8, -6), runif(1, -6, -4),
runif(1, -10, -8), runif(1, -8, -6), runif(1, -6, -4)),
"Upper_CI" = c(runif(1, 8, 10), runif(1, 6, 8), runif(1, 4, 6),
runif(1, 8, 10), runif(1, 6, 8), runif(1, 4, 6),
runif(1, 8, 10), runif(1, 6, 8), runif(1, 4, 6),
runif(1, 8, 10), runif(1, 6, 8), runif(1, 4, 6),
runif(1, 8, 10), runif(1, 6, 8), runif(1, 4, 6),
runif(1, 8, 10), runif(1, 6, 8), runif(1, 4, 6),
runif(1, 8, 10), runif(1, 6, 8), runif(1, 4, 6),
runif(1, 8, 10), runif(1, 6, 8), runif(1, 4, 6)),
"Species" = factor(c(rep("A", 12), rep("B", 12))),
"Parameter" = factor(paste0("P", c(rep(1,3), rep(2,3), rep(5,3),
rep(3,3), rep(4,3), rep(6,3),
rep(7,3), rep(8,3)))))
ggplot(dat, aes(x = as.integer(Parameter))) +
geom_rect(aes(xmin = as.integer(Parameter) - 0.5,
xmax = as.integer(Parameter) + 0.5,
ymin = Lower_CI,
ymax = Upper_CI,
fill = as.integer(CredInt))) +
scale_x_continuous(breaks = 1:length(levels(dat$Parameter)),labels = levels(dat$Parameter)) +
facet_wrap(~Species, scales = "free_x")