Hi,
I'm trying to create a Boxplot with lines between dots of 2 conditions (PRE & POS) from an excel table. However, in this excel table, some rows are empty (cause of lack of data). I guess this is posing a problem for establishing my lines between my 2 conditions. Here is the script I used (working when I have fully filled excel rows):
data<-read_excel("Thelper PRE POS.xls")
Samples<-data$samples
Samples=as.factor(data$samples)
Samples
Th1<-data$Th1
condi<-data$condi
Condi=as.factor(data$condi)
Condi<-factor(c("pre","pos"))
dfdata<-data.frame(condi,Th1)
dfdata<-drop_na(dfdata)
p<-ggplot(dfdata,aes(x=reorder(condi,Th1),y=Th1))+
geom_boxplot(aes(fill=condi),width=0.4, size=1,fatten=0.3,colour="black")+
geom_point(size=2, colour="black")+
geom_line(aes(group=Samples),colour="grey",linetype="11")+
scale_fill_manual(values = c("#C3D7A4", "#CC79A7"))+
theme_classic()
p
Here is the error I get:
geom_path: Each group consists of only one observation. Do you need to adjust the group
aesthetic?
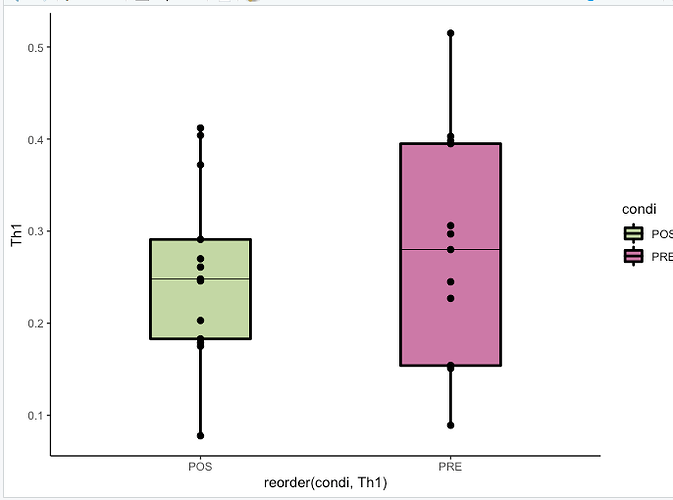
And lines are not present on my graphs:
What can be the problem?
Thanks a lot