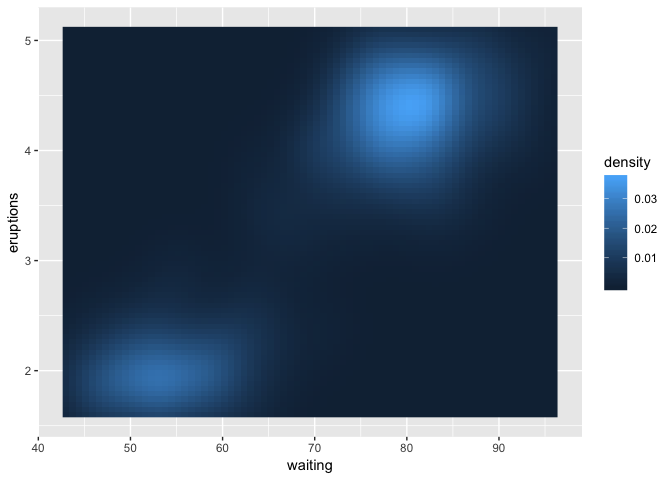
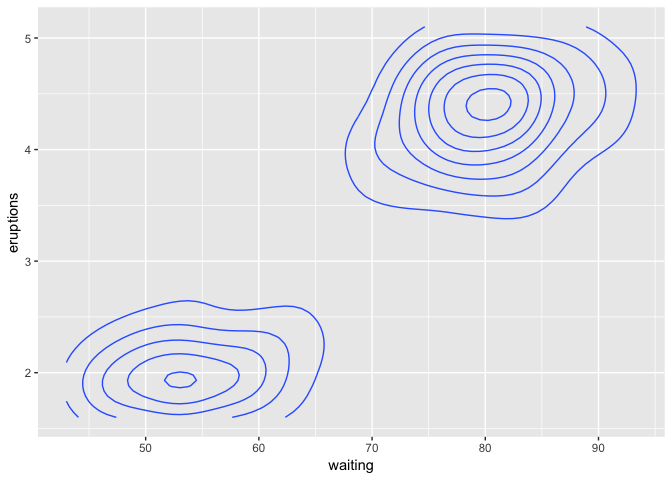
It depends on the data. For the data above, not really in any useful way, as there's just not enough of it. If you have more and it's 2-dimensional, that's what geom_density_2d implicitly does. If your data is on an easily-completed grid, e.g. integers, you can use tidyr::complete with fill = list(z = 0), but this is making some assumptions about your data that may not be true.
Another option is to regress z on x and y with a model—maybe a flexible one like LOESS or splines, possibly with constraints—and then predict on a grid of x and y values. This approach could generate a result out of anything, but may require a lot of tweaking depending on what you're after.
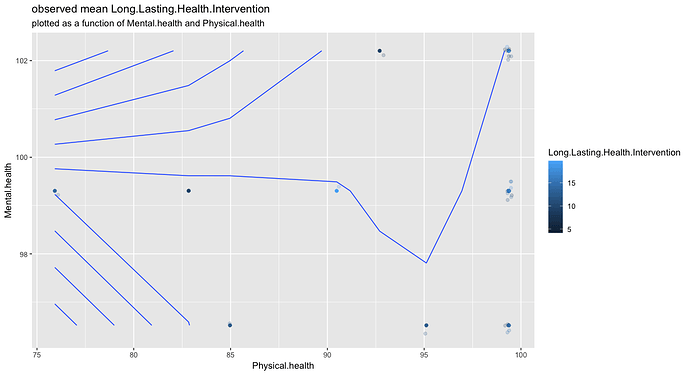
A very simple example using your three points and lm:
library(ggplot2)
df <- data.frame(x = c(7L, 1L, 5L),
y = c(6, 3.14, 2.7),
z = c(5.3, 4, 6.1))
grid <- expand.grid(x = seq(10), y = seq(10))
grid$z <- predict(lm(z ~ ., df), grid)
ggplot(grid, aes(x, y, z = z)) + geom_contour()
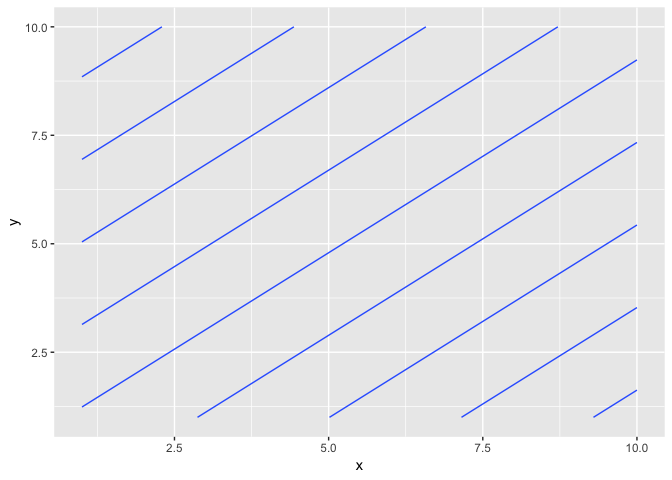
...but this generates lines. If you look at the raster plot you can see why:
ggplot(grid, aes(x, y, fill = z)) + geom_raster()
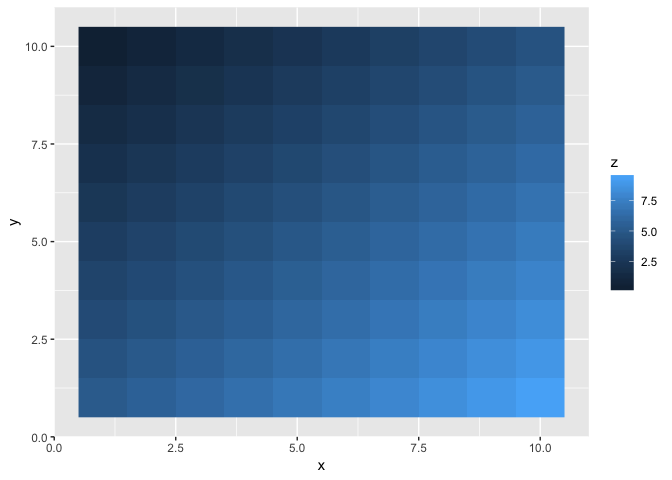
It generated a plane passing through the three points, which is what lm does.