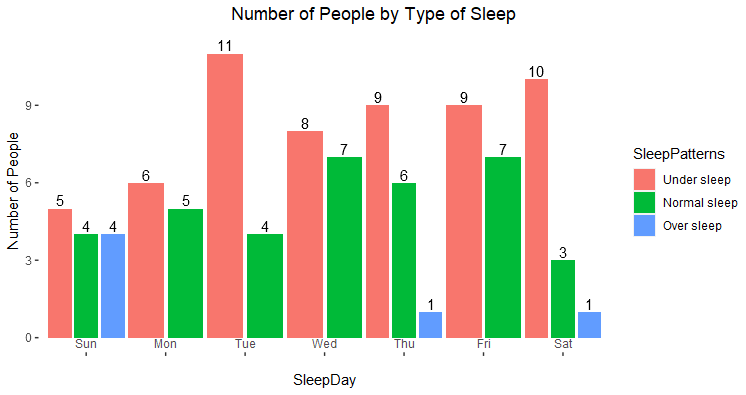
Here is a suggestion
library(tidyverse)
library(ggrepel)
mydata <- structure(list(Id = c(
1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1644430081, 1644430081, 1644430081, 1927972279,
1927972279, 1927972279, 1927972279, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2320127002, 2347167796, 2347167796, 2347167796, 2347167796,
2347167796, 2347167796, 2347167796, 2347167796, 2347167796, 2347167796,
2347167796, 2347167796, 2347167796, 2347167796, 2347167796, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714
), SleepDay = structure(c(
3L, 4L, 6L, 7L, 3L, 4L, 5L, 7L, 1L,
2L, 3L, 5L, 6L, 7L, 1L, 2L, 3L, 5L, 6L, 7L, 1L, 2L, 3L, 4L, 6L,
7L, 1L, 4L, 6L, 3L, 5L, 3L, 4L, 5L, 6L, 7L, 1L, 3L, 4L, 5L, 6L,
7L, 1L, 2L, 4L, 5L, 6L, 7L, 1L, 2L, 4L, 5L, 6L, 7L, 1L, 2L, 3L,
4L, 5L, 7L, 4L, 5L, 6L, 1L, 2L, 3L, 5L, 6L, 7L, 1L, 2L, 3L, 4L,
5L, 6L, 3L, 4L, 5L, 6L, 7L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 1L, 2L,
3L, 4L, 5L, 6L, 7L, 1L, 2L, 3L, 4L, 5L, 6L
), levels = c(
"Sun",
"Mon", "Tue", "Wed", "Thu", "Fri", "Sat"
), class = c(
"ordered",
"factor"
)), TotalSleepRecords = c(
1L, 2L, 1L, 2L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L
), TotalMinutesAsleep = c(
327L,
384L, 412L, 340L, 304L, 360L, 325L, 361L, 430L, 277L, 245L, 366L,
341L, 404L, 369L, 277L, 273L, 247L, 334L, 331L, 594L, 338L, 383L,
285L, 119L, 124L, 137L, 398L, 475L, 296L, 166L, 503L, 531L, 545L,
523L, 524L, 437L, 498L, 461L, 477L, 520L, 522L, 555L, 506L, 508L,
513L, 490L, 573L, 527L, 511L, 538L, 468L, 524L, 511L, 541L, 531L,
357L, 523L, 456L, 61L, 467L, 445L, 452L, 556L, 500L, 465L, 460L,
405L, 374L, 442L, 433L, 436L, 448L, 408L, 411L, 274L, 295L, 291L,
424L, 283L, 381L, 412L, 219L, 152L, 332L, 355L, 235L, 310L, 262L,
250L, 349L, 261L, 333L, 237L, 383L, 230L, 292L, 213L, 318L, 323L
), TotalTimeInBed = c(
346L, 407L, 442L, 367L, 320L, 377L, 364L,
384L, 449L, 323L, 274L, 393L, 354L, 425L, 396L, 309L, 296L, 264L,
367L, 349L, 611L, 342L, 403L, 306L, 127L, 142L, 154L, 422L, 499L,
315L, 178L, 546L, 565L, 568L, 573L, 567L, 498L, 540L, 510L, 514L,
545L, 554L, 591L, 531L, 545L, 545L, 510L, 607L, 546L, 543L, 560L,
485L, 548L, 521L, 568L, 556L, 380L, 553L, 485L, 69L, 531L, 489L,
504L, 602L, 557L, 514L, 484L, 461L, 386L, 459L, 471L, 490L, 499L,
450L, 473L, 469L, 456L, 397L, 556L, 510L, 566L, 522L, 395L, 305L,
512L, 476L, 372L, 526L, 467L, 371L, 540L, 423L, 478L, 382L, 626L,
384L, 500L, 336L, 480L, 512L
), SleepPatterns = structure(c(
1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 3L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 2L,
3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 2L, 2L, 2L, 2L, 3L, 2L,
2L, 2L, 2L, 2L, 2L, 3L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 3L, 2L,
2L, 2L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L
), levels = c("Under sleep", "Normal sleep", "Over sleep"), class = "factor"), TotalSittingTime = c(
19L, 23L, 30L, 27L,
16L, 17L, 39L, 23L, 19L, 46L, 29L, 27L, 13L, 21L, 27L, 32L, 23L,
17L, 33L, 18L, 17L, 4L, 20L, 21L, 8L, 18L, 17L, 24L, 24L, 19L,
12L, 43L, 34L, 23L, 50L, 43L, 61L, 42L, 49L, 37L, 25L, 32L, 36L,
25L, 37L, 32L, 20L, 34L, 19L, 32L, 22L, 17L, 24L, 10L, 27L, 25L,
23L, 30L, 29L, 8L, 64L, 44L, 52L, 46L, 57L, 49L, 24L, 56L, 12L,
17L, 38L, 54L, 51L, 42L, 62L, 195L, 161L, 106L, 132L, 227L, 185L,
110L, 176L, 153L, 180L, 121L, 137L, 216L, 205L, 121L, 191L, 162L,
145L, 145L, 243L, 154L, 208L, 123L, 162L, 189L
)), row.names = c(
NA,
100L
), class = "data.frame")
mydata_smry <- mydata |>
group_by(SleepDay, SleepPatterns) |>
summarise(n = n())
ggplot(mydata_smry, aes(x = SleepDay, fill = SleepPatterns)) +
geom_col(aes(y=n),
position = position_dodge(width = 1)) +
theme(panel.background = element_blank()) +
theme(plot.background = element_blank()) +
theme(
plot.title.position = "plot",
plot.title = element_text(hjust = 0.5)
) +
theme(axis.text.x = element_text(vjust = 10)) +
ggtitle("Number of People by Type of Sleep") +
ylab("Number of People") +
geom_text(
aes(
label = n,
y = n + .3
),
position = position_dodge(width = 1)
)