Hi,
I work on the public dataset Bellabeat and I want to plot a geom_bar with SleepPatterns (Categorical variable) by week day (i.e. Monday, Tuesday etc).
ggplot(sleepday_daily, aes(x = SleepDay , fill = factor(SleepPatterns))) +
geom_bar(position = "dodge")+
theme(panel.background = element_blank())+
theme(plot.background = element_blank())+
theme(plot.title.position = 'plot',
plot.title = element_text(hjust = 0.5))+
theme(axis.text.x = element_text(vjust = 10))+
ggtitle("Number of People by Type of Sleep")+
ylab("Number of People")+
stat_count(geom = "text",
aes(label = stat(count)),
position=position_fill(vjust=30), colour="black")
Sleepday column is Monday, Tuesday etc and SleepPatterns UnderSleep,Normal Sleep and Oversleep.
I get label values but there is overlapping (one value on the top of the other )
Any tip what I can do? I have googled everything
Thanks
Panos
Thanks for the code but could you supply us with some sample data as well. A handy way to supply some sample data is the dput() function. In the case of a large dataset something like dput(head(mydata, 100)) should supply the data we need. Just do dput(mydata) where mydata is your data. Copy the output and paste it here.
By my count the dataset Bellabeat contains 18 .csv files and it could be a mess trying to figure out exactly what your working dataset looks like,
Thank you @jrkrideau
structure(list(Id = c(1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1644430081, 1644430081, 1644430081, 1927972279,
1927972279, 1927972279, 1927972279, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2320127002, 2347167796, 2347167796, 2347167796, 2347167796,
2347167796, 2347167796, 2347167796, 2347167796, 2347167796, 2347167796,
2347167796, 2347167796, 2347167796, 2347167796, 2347167796, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714
), SleepDay = structure(c(3L, 4L, 6L, 7L, 3L, 4L, 5L, 7L, 1L,
2L, 3L, 5L, 6L, 7L, 1L, 2L, 3L, 5L, 6L, 7L, 1L, 2L, 3L, 4L, 6L,
7L, 1L, 4L, 6L, 3L, 5L, 3L, 4L, 5L, 6L, 7L, 1L, 3L, 4L, 5L, 6L,
7L, 1L, 2L, 4L, 5L, 6L, 7L, 1L, 2L, 4L, 5L, 6L, 7L, 1L, 2L, 3L,
4L, 5L, 7L, 4L, 5L, 6L, 1L, 2L, 3L, 5L, 6L, 7L, 1L, 2L, 3L, 4L,
5L, 6L, 3L, 4L, 5L, 6L, 7L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 1L, 2L,
3L, 4L, 5L, 6L, 7L, 1L, 2L, 3L, 4L, 5L, 6L), levels = c("Sun",
"Mon", "Tue", "Wed", "Thu", "Fri", "Sat"), class = c("ordered",
"factor")), TotalSleepRecords = c(1L, 2L, 1L, 2L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L), TotalMinutesAsleep = c(327L,
384L, 412L, 340L, 304L, 360L, 325L, 361L, 430L, 277L, 245L, 366L,
341L, 404L, 369L, 277L, 273L, 247L, 334L, 331L, 594L, 338L, 383L,
285L, 119L, 124L, 137L, 398L, 475L, 296L, 166L, 503L, 531L, 545L,
523L, 524L, 437L, 498L, 461L, 477L, 520L, 522L, 555L, 506L, 508L,
513L, 490L, 573L, 527L, 511L, 538L, 468L, 524L, 511L, 541L, 531L,
357L, 523L, 456L, 61L, 467L, 445L, 452L, 556L, 500L, 465L, 460L,
405L, 374L, 442L, 433L, 436L, 448L, 408L, 411L, 274L, 295L, 291L,
424L, 283L, 381L, 412L, 219L, 152L, 332L, 355L, 235L, 310L, 262L,
250L, 349L, 261L, 333L, 237L, 383L, 230L, 292L, 213L, 318L, 323L
), TotalTimeInBed = c(346L, 407L, 442L, 367L, 320L, 377L, 364L,
384L, 449L, 323L, 274L, 393L, 354L, 425L, 396L, 309L, 296L, 264L,
367L, 349L, 611L, 342L, 403L, 306L, 127L, 142L, 154L, 422L, 499L,
315L, 178L, 546L, 565L, 568L, 573L, 567L, 498L, 540L, 510L, 514L,
545L, 554L, 591L, 531L, 545L, 545L, 510L, 607L, 546L, 543L, 560L,
485L, 548L, 521L, 568L, 556L, 380L, 553L, 485L, 69L, 531L, 489L,
504L, 602L, 557L, 514L, 484L, 461L, 386L, 459L, 471L, 490L, 499L,
450L, 473L, 469L, 456L, 397L, 556L, 510L, 566L, 522L, 395L, 305L,
512L, 476L, 372L, 526L, 467L, 371L, 540L, 423L, 478L, 382L, 626L,
384L, 500L, 336L, 480L, 512L), SleepPatterns = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 3L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 2L,
3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 2L, 2L, 2L, 2L, 3L, 2L,
2L, 2L, 2L, 2L, 2L, 3L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 3L, 2L,
2L, 2L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L), levels = c("Under sleep", "Normal sleep", "Over sleep"
), class = "factor"), TotalSittingTime = c(19L, 23L, 30L, 27L,
16L, 17L, 39L, 23L, 19L, 46L, 29L, 27L, 13L, 21L, 27L, 32L, 23L,
17L, 33L, 18L, 17L, 4L, 20L, 21L, 8L, 18L, 17L, 24L, 24L, 19L,
12L, 43L, 34L, 23L, 50L, 43L, 61L, 42L, 49L, 37L, 25L, 32L, 36L,
25L, 37L, 32L, 20L, 34L, 19L, 32L, 22L, 17L, 24L, 10L, 27L, 25L,
23L, 30L, 29L, 8L, 64L, 44L, 52L, 46L, 57L, 49L, 24L, 56L, 12L,
17L, 38L, 54L, 51L, 42L, 62L, 195L, 161L, 106L, 132L, 227L, 185L,
110L, 176L, 153L, 180L, 121L, 137L, 216L, 205L, 121L, 191L, 162L,
145L, 145L, 243L, 154L, 208L, 123L, 162L, 189L)), row.names = c(NA,
100L), class = "data.frame")
dput(mydata) output (I removed 100)
structure(list(Id = c(1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366), SleepDay = structure(c(3L, 4L, 6L, 7L,
3L, 4L), levels = c("Sun", "Mon", "Tue", "Wed", "Thu", "Fri",
"Sat"), class = c("ordered", "factor")), TotalSleepRecords = c(1L,
2L, 1L, 2L, 1L, 1L), TotalMinutesAsleep = c(327L, 384L, 412L,
340L, 304L, 360L), TotalTimeInBed = c(346L, 407L, 442L, 367L,
320L, 377L), SleepPatterns = structure(c(1L, 1L, 1L, 1L, 1L,
1L), levels = c("Under sleep", "Normal sleep", "Over sleep"), class = "factor"),
TotalSittingTime = c(19L, 23L, 30L, 27L, 16L, 17L)), row.names = c(NA,
6L), class = "data.frame")
I think I see what is happening but at the moment I don't see how the cure it except by brute fore and trial and error.
What I think is happening is that the stat_count() function is counting in integers and getting 17 integers that it then that it then uses to place the numbers. The problem in that the x-asis is only 7 units long and the bar spacing is irregular. Some days have 2 bars and some have three.
I tried by a different method and got the some result except that I also messed up the order of tho day.
I'll keep at it but I think we need a real expert.
Do you REALLY need those numbers? People like Tufte would call them chart junk.
Here is a suggestion
library(tidyverse)
library(ggrepel)
mydata <- structure(list(Id = c(
1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1503960366, 1503960366, 1503960366, 1503960366,
1503960366, 1503960366, 1644430081, 1644430081, 1644430081, 1927972279,
1927972279, 1927972279, 1927972279, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2026352035, 2026352035, 2026352035, 2026352035, 2026352035,
2026352035, 2320127002, 2347167796, 2347167796, 2347167796, 2347167796,
2347167796, 2347167796, 2347167796, 2347167796, 2347167796, 2347167796,
2347167796, 2347167796, 2347167796, 2347167796, 2347167796, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714,
3977333714, 3977333714, 3977333714, 3977333714, 3977333714, 3977333714
), SleepDay = structure(c(
3L, 4L, 6L, 7L, 3L, 4L, 5L, 7L, 1L,
2L, 3L, 5L, 6L, 7L, 1L, 2L, 3L, 5L, 6L, 7L, 1L, 2L, 3L, 4L, 6L,
7L, 1L, 4L, 6L, 3L, 5L, 3L, 4L, 5L, 6L, 7L, 1L, 3L, 4L, 5L, 6L,
7L, 1L, 2L, 4L, 5L, 6L, 7L, 1L, 2L, 4L, 5L, 6L, 7L, 1L, 2L, 3L,
4L, 5L, 7L, 4L, 5L, 6L, 1L, 2L, 3L, 5L, 6L, 7L, 1L, 2L, 3L, 4L,
5L, 6L, 3L, 4L, 5L, 6L, 7L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 1L, 2L,
3L, 4L, 5L, 6L, 7L, 1L, 2L, 3L, 4L, 5L, 6L
), levels = c(
"Sun",
"Mon", "Tue", "Wed", "Thu", "Fri", "Sat"
), class = c(
"ordered",
"factor"
)), TotalSleepRecords = c(
1L, 2L, 1L, 2L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L
), TotalMinutesAsleep = c(
327L,
384L, 412L, 340L, 304L, 360L, 325L, 361L, 430L, 277L, 245L, 366L,
341L, 404L, 369L, 277L, 273L, 247L, 334L, 331L, 594L, 338L, 383L,
285L, 119L, 124L, 137L, 398L, 475L, 296L, 166L, 503L, 531L, 545L,
523L, 524L, 437L, 498L, 461L, 477L, 520L, 522L, 555L, 506L, 508L,
513L, 490L, 573L, 527L, 511L, 538L, 468L, 524L, 511L, 541L, 531L,
357L, 523L, 456L, 61L, 467L, 445L, 452L, 556L, 500L, 465L, 460L,
405L, 374L, 442L, 433L, 436L, 448L, 408L, 411L, 274L, 295L, 291L,
424L, 283L, 381L, 412L, 219L, 152L, 332L, 355L, 235L, 310L, 262L,
250L, 349L, 261L, 333L, 237L, 383L, 230L, 292L, 213L, 318L, 323L
), TotalTimeInBed = c(
346L, 407L, 442L, 367L, 320L, 377L, 364L,
384L, 449L, 323L, 274L, 393L, 354L, 425L, 396L, 309L, 296L, 264L,
367L, 349L, 611L, 342L, 403L, 306L, 127L, 142L, 154L, 422L, 499L,
315L, 178L, 546L, 565L, 568L, 573L, 567L, 498L, 540L, 510L, 514L,
545L, 554L, 591L, 531L, 545L, 545L, 510L, 607L, 546L, 543L, 560L,
485L, 548L, 521L, 568L, 556L, 380L, 553L, 485L, 69L, 531L, 489L,
504L, 602L, 557L, 514L, 484L, 461L, 386L, 459L, 471L, 490L, 499L,
450L, 473L, 469L, 456L, 397L, 556L, 510L, 566L, 522L, 395L, 305L,
512L, 476L, 372L, 526L, 467L, 371L, 540L, 423L, 478L, 382L, 626L,
384L, 500L, 336L, 480L, 512L
), SleepPatterns = structure(c(
1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 3L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 2L,
3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 2L, 2L, 2L, 2L, 3L, 2L,
2L, 2L, 2L, 2L, 2L, 3L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 3L, 2L,
2L, 2L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L
), levels = c("Under sleep", "Normal sleep", "Over sleep"), class = "factor"), TotalSittingTime = c(
19L, 23L, 30L, 27L,
16L, 17L, 39L, 23L, 19L, 46L, 29L, 27L, 13L, 21L, 27L, 32L, 23L,
17L, 33L, 18L, 17L, 4L, 20L, 21L, 8L, 18L, 17L, 24L, 24L, 19L,
12L, 43L, 34L, 23L, 50L, 43L, 61L, 42L, 49L, 37L, 25L, 32L, 36L,
25L, 37L, 32L, 20L, 34L, 19L, 32L, 22L, 17L, 24L, 10L, 27L, 25L,
23L, 30L, 29L, 8L, 64L, 44L, 52L, 46L, 57L, 49L, 24L, 56L, 12L,
17L, 38L, 54L, 51L, 42L, 62L, 195L, 161L, 106L, 132L, 227L, 185L,
110L, 176L, 153L, 180L, 121L, 137L, 216L, 205L, 121L, 191L, 162L,
145L, 145L, 243L, 154L, 208L, 123L, 162L, 189L
)), row.names = c(
NA,
100L
), class = "data.frame")
mydata_smry <- mydata |>
group_by(SleepDay, SleepPatterns) |>
summarise(n = n())
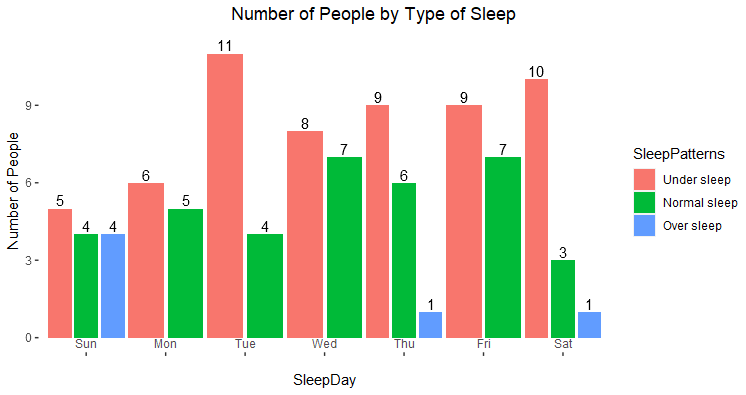
ggplot(mydata_smry, aes(x = SleepDay, fill = SleepPatterns)) +
geom_col(aes(y=n),
position = position_dodge(width = 1)) +
theme(panel.background = element_blank()) +
theme(plot.background = element_blank()) +
theme(
plot.title.position = "plot",
plot.title = element_text(hjust = 0.5)
) +
theme(axis.text.x = element_text(vjust = 10)) +
ggtitle("Number of People by Type of Sleep") +
ylab("Number of People") +
geom_text(
aes(
label = n,
y = n + .3
),
position = position_dodge(width = 1)
)
@nirgrahamuk : Thank you very much! you are a star!
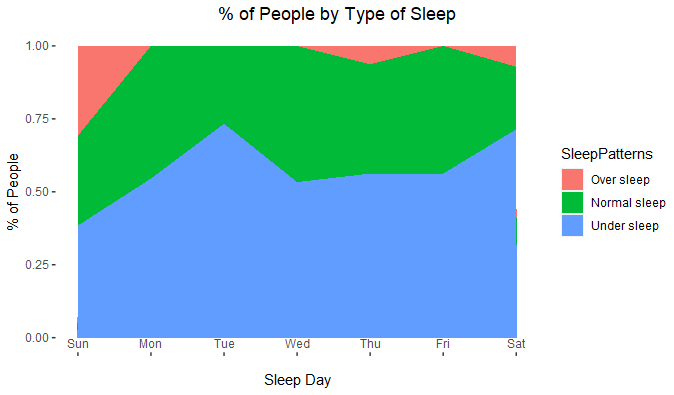
@jrkrideau : Many thanks for your feedback. The aim of that graph was to show sleep patterns by day. I think that over sleep patterns should be removed as it is not shown all days and it looks as an outlier. How would you present this info?
If I look at nirgrahamuk's plot and assume a Western European or North American workweek I think you would be losing useful data.
I mean, the Saturday & Sunday oversleep is fairly explainable but why Thursday? I'd definitively leave Oversleep in.
Do you have duration of Oversleep? It might be interesting to plot that. Or season of the year? I am in Canada where sleep patterns are very affected by the season.
It really depends on the research question.
mydata_smry1 <- mydata |>
group_by(SleepDay, SleepPatterns) |>
summarise(n = n()) |> ungroup() |>
tidyr::complete(SleepDay,
SleepPatterns,
fill = list(n=0L))
mydata_smry2 <- mydata |>
group_by(SleepDay) |>
summarise(total = n())
mydata_smry3 <- left_join(
mydata_smry2,
mydata_smry1
) |> mutate(
SleepPatterns = forcats::fct_rev(SleepPatterns),
pcnt =n / total
)
ggplot(mydata_smry3, aes(x = as.numeric(SleepDay),
y=pcnt
,fill = SleepPatterns)
) +
geom_area(position=position_stack()) +
theme(panel.background = element_blank()) +
theme(plot.background = element_blank()) +
theme(
plot.title.position = "plot",
plot.title = element_text(hjust = 0.5)
) +
theme(axis.text.x = element_text(vjust = 10)) +
ggtitle("% of People by Type of Sleep") +
ylab("% of People") +
scale_x_continuous(breaks = 1:length(levels(mydata_smry3$SleepDay)), labels = levels(mydata_smry3$SleepDay),
name = "Sleep Day")
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.