Hi, I have a question how to make a "normal" table from crosstab 2x2 or 3x3 contingency table ?
I mean I do not want summary table but a table with individual values.
Hi,
It is not immediately clear how you want us to help. It always helps to add a reprex (see here: FAQ: How to do a minimal reproducible example ( reprex ) for beginners ).
You will see below that I have created a base R way and melt way to take a table and convert it into a list of values. Let me know if this is what you wanted?
# load table
crosstable <- structure(list(col1 = c(0, 0, 1, 0, 0, 0, 0, 0, 0), col2 = c(0, 1, 0, 0, 0, 0, 0, 0, 0), col3 = c(0, 0, 0, 0, 0, 0, 0, 0, 1), col4 = c(1, 0, 0, 0, 0, 0, 0, 0, 0), col5 = c(0, 0, 0, 0, 0, 0, 0, 1, 0), col6 = c(0, 0, 0, 0, 0, 1, 0, 0, 0), col7 = c(0, 0, 0, 0, 1, 0, 0, 0, 0), col8 = c(0, 0, 0, 0, 0, 0, 1, 0, 0)), .Names = c("col1", "col2", "col3", "col4", "col5", "col6", "col7", "col8"), row.names = c("row1", "row2", "row3", "row4", "row5", "row6", "row7", "row8", "row9"), class = "data.frame")
#display table
crosstable
#> col1 col2 col3 col4 col5 col6 col7 col8
#> row1 0 0 0 1 0 0 0 0
#> row2 0 1 0 0 0 0 0 0
#> row3 1 0 0 0 0 0 0 0
#> row4 0 0 0 0 0 0 0 0
#> row5 0 0 0 0 0 0 1 0
#> row6 0 0 0 0 0 1 0 0
#> row7 0 0 0 0 0 0 0 1
#> row8 0 0 0 0 1 0 0 0
#> row9 0 0 1 0 0 0 0 0
CTLong <- data.frame(rows = rownames(crosstable), stack(crosstable))
CTLong <- CTLong[order(CTLong$rows), ]
CTLong
#> rows values ind
#> 1 row1 0 col1
#> 10 row1 0 col2
#> 19 row1 0 col3
#> 28 row1 1 col4
#> 37 row1 0 col5
#> 46 row1 0 col6
#> 55 row1 0 col7
#> 64 row1 0 col8
#> 2 row2 0 col1
#> 11 row2 1 col2
#> 20 row2 0 col3
#> 29 row2 0 col4
#> 38 row2 0 col5
#> 47 row2 0 col6
#> 56 row2 0 col7
#> 65 row2 0 col8
#> 3 row3 1 col1
#> 12 row3 0 col2
#> 21 row3 0 col3
#> 30 row3 0 col4
#> 39 row3 0 col5
#> 48 row3 0 col6
#> 57 row3 0 col7
#> 66 row3 0 col8
#> 4 row4 0 col1
#> 13 row4 0 col2
#> 22 row4 0 col3
#> 31 row4 0 col4
#> 40 row4 0 col5
#> 49 row4 0 col6
#> 58 row4 0 col7
#> 67 row4 0 col8
#> 5 row5 0 col1
#> 14 row5 0 col2
#> 23 row5 0 col3
#> 32 row5 0 col4
#> 41 row5 0 col5
#> 50 row5 0 col6
#> 59 row5 1 col7
#> 68 row5 0 col8
#> 6 row6 0 col1
#> 15 row6 0 col2
#> 24 row6 0 col3
#> 33 row6 0 col4
#> 42 row6 0 col5
#> 51 row6 1 col6
#> 60 row6 0 col7
#> 69 row6 0 col8
#> 7 row7 0 col1
#> 16 row7 0 col2
#> 25 row7 0 col3
#> 34 row7 0 col4
#> 43 row7 0 col5
#> 52 row7 0 col6
#> 61 row7 0 col7
#> 70 row7 1 col8
#> 8 row8 0 col1
#> 17 row8 0 col2
#> 26 row8 0 col3
#> 35 row8 0 col4
#> 44 row8 1 col5
#> 53 row8 0 col6
#> 62 row8 0 col7
#> 71 row8 0 col8
#> 9 row9 0 col1
#> 18 row9 0 col2
#> 27 row9 1 col3
#> 36 row9 0 col4
#> 45 row9 0 col5
#> 54 row9 0 col6
#> 63 row9 0 col7
#> 72 row9 0 col8
library(reshape2)
CTLong2 <- melt(cbind(rownames(crosstable),crosstable))
#> Using rownames(crosstable) as id variables
CTLong2
#> rownames(crosstable) variable value
#> 1 row1 col1 0
#> 2 row2 col1 0
#> 3 row3 col1 1
#> 4 row4 col1 0
#> 5 row5 col1 0
#> 6 row6 col1 0
#> 7 row7 col1 0
#> 8 row8 col1 0
#> 9 row9 col1 0
#> 10 row1 col2 0
#> 11 row2 col2 1
#> 12 row3 col2 0
#> 13 row4 col2 0
#> 14 row5 col2 0
#> 15 row6 col2 0
#> 16 row7 col2 0
#> 17 row8 col2 0
#> 18 row9 col2 0
#> 19 row1 col3 0
#> 20 row2 col3 0
#> 21 row3 col3 0
#> 22 row4 col3 0
#> 23 row5 col3 0
#> 24 row6 col3 0
#> 25 row7 col3 0
#> 26 row8 col3 0
#> 27 row9 col3 1
#> 28 row1 col4 1
#> 29 row2 col4 0
#> 30 row3 col4 0
#> 31 row4 col4 0
#> 32 row5 col4 0
#> 33 row6 col4 0
#> 34 row7 col4 0
#> 35 row8 col4 0
#> 36 row9 col4 0
#> 37 row1 col5 0
#> 38 row2 col5 0
#> 39 row3 col5 0
#> 40 row4 col5 0
#> 41 row5 col5 0
#> 42 row6 col5 0
#> 43 row7 col5 0
#> 44 row8 col5 1
#> 45 row9 col5 0
#> 46 row1 col6 0
#> 47 row2 col6 0
#> 48 row3 col6 0
#> 49 row4 col6 0
#> 50 row5 col6 0
#> 51 row6 col6 1
#> 52 row7 col6 0
#> 53 row8 col6 0
#> 54 row9 col6 0
#> 55 row1 col7 0
#> 56 row2 col7 0
#> 57 row3 col7 0
#> 58 row4 col7 0
#> 59 row5 col7 1
#> 60 row6 col7 0
#> 61 row7 col7 0
#> 62 row8 col7 0
#> 63 row9 col7 0
#> 64 row1 col8 0
#> 65 row2 col8 0
#> 66 row3 col8 0
#> 67 row4 col8 0
#> 68 row5 col8 0
#> 69 row6 col8 0
#> 70 row7 col8 1
#> 71 row8 col8 0
#> 72 row9 col8 0
Created on 2021-11-07 by the reprex package (v2.0.0)
Thank you, this is probably not what I want, I would like to achieve this:
my_datas2 <- tribble(~Answers, ~Group_1, ~Group_2,
'A', 8, 4,
'B', 6, 7,
'C', 8, 9)
#or
my_datas3 <- tribble(~Answers, ~Group, ~Values,
"A", "Group_1", 8,
"B", "Group_1", 6,
"C", "Group_1", 8,
"A", "Group_2", 4,
"B", "Group_2", 7,
"C", "Group_2", 9)
desired result:
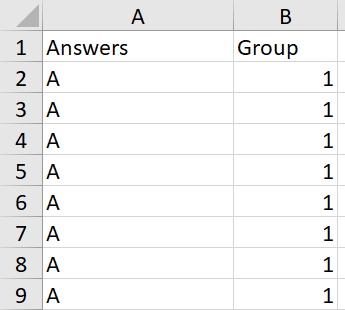
and so on with replies B and C continuing down in the table for Group 2 as well.
additionally if you could help me with transforming my_datas2 to my_datas3 as here I have done it manually by re-writing it. Maybe there is a code for that. Thank you.
Here's a tidyr based solution:
my_datas2 <- tibble::tribble(~Answers, ~Group_1, ~Group_2,
'A', 8, 4,
'B', 6, 7,
'C', 8, 9)
tidyr::pivot_longer(data = my_datas2,
cols = -Answers,
names_to = "Group",
values_to = "Values")
#> # A tibble: 6 × 3
#> Answers Group Values
#> <chr> <chr> <dbl>
#> 1 A Group_1 8
#> 2 A Group_2 4
#> 3 B Group_1 6
#> 4 B Group_2 7
#> 5 C Group_1 8
#> 6 C Group_2 9
Hope this helps.
Hi there,
See my example below to take yourmy_datas2 and converting it into the desired result. You can just do some text clean up on the Group column but I kept as is so you can see it is working. Definitely have a look at how @Yarnabrina did the pivot_longer it is smiliar to gather but I think that is the current correct/most recent way of doing it.
library(tibble)
#> Warning: package 'tibble' was built under R version 4.0.5
library(tidyr)
#> Warning: package 'tidyr' was built under R version 4.0.5
#creating your data
my_datas2 <- tibble(
answers = c("A","B","C"),
group_1 = c(8,6,8),
group_2 = c(4,7,9)
)
my_datas2
#> # A tibble: 3 x 3
#> answers group_1 group_2
#> <chr> <dbl> <dbl>
#> 1 A 8 4
#> 2 B 6 7
#> 3 C 8 9
#converting your data from wide to long
dat <-
my_datas2 %>% gather("my_datas2", "value", -answers)
dat
#> # A tibble: 6 x 3
#> answers my_datas2 value
#> <chr> <chr> <dbl>
#> 1 A group_1 8
#> 2 B group_1 6
#> 3 C group_1 8
#> 4 A group_2 4
#> 5 B group_2 7
#> 6 C group_2 9
#creating the repition based on the original cell values
test <- data.frame(
Answers = do.call("c", (mapply(rep, dat$answers, dat$value))),
Group = do.call("c", (mapply(rep, dat$my_datas2, dat$value)))
)
test
#> Answers Group
#> 1 A group_1
#> 2 A group_1
#> 3 A group_1
#> 4 A group_1
#> 5 A group_1
#> 6 A group_1
#> 7 A group_1
#> 8 A group_1
#> 9 B group_1
#> 10 B group_1
#> 11 B group_1
#> 12 B group_1
#> 13 B group_1
#> 14 B group_1
#> 15 C group_1
#> 16 C group_1
#> 17 C group_1
#> 18 C group_1
#> 19 C group_1
#> 20 C group_1
#> 21 C group_1
#> 22 C group_1
#> 23 A group_2
#> 24 A group_2
#> 25 A group_2
#> 26 A group_2
#> 27 B group_2
#> 28 B group_2
#> 29 B group_2
#> 30 B group_2
#> 31 B group_2
#> 32 B group_2
#> 33 B group_2
#> 34 C group_2
#> 35 C group_2
#> 36 C group_2
#> 37 C group_2
#> 38 C group_2
#> 39 C group_2
#> 40 C group_2
#> 41 C group_2
#> 42 C group_2
Created on 2021-11-07 by the reprex package (v2.0.0)
Thank you @Yarnabrina for your kind reply and thank you @GreyMerchant for your fantastic solution,
This is what I wanted.
Could you please explain a bit what does this code do:
test <- data.frame(
Answers = do.call("c", (mapply(rep, dat$answers, dat$value))),
Group = do.call("c", (mapply(rep, dat$my_datas2, dat$value)))
)
Based on @GreyMerchant's answer and it's acceptance by @Andrzej, I clearly misunderstood the requirement. In that case, please feel free to point that out, liking a wrong reply creates possible confusion for future visitors.
Anyway, I'll let @GreyMerchant explain his solution. Here are two more solutions, one's continuing my prevous reply, and one's a modification of @GreyMerchant's do.call use to make it a bit short:
my_datas2 <- tibble::tribble(~Answers, ~Group_1, ~Group_2,
'A', 8, 4,
'B', 6, 7,
'C', 8, 9)
# continuing previous solution
my_datas2 |>
tidyr::pivot_longer(cols = -Answers,
names_to = "Group",
values_to = "Values") |>
tidyr::uncount(weights = Values,
.remove = TRUE) |>
as.data.frame()
#> Answers Group
#> 1 A Group_1
#> 2 A Group_1
#> 3 A Group_1
#> 4 A Group_1
#> 5 A Group_1
#> 6 A Group_1
#> 7 A Group_1
#> 8 A Group_1
#> 9 A Group_2
#> 10 A Group_2
#> 11 A Group_2
#> 12 A Group_2
#> 13 B Group_1
#> 14 B Group_1
#> 15 B Group_1
#> 16 B Group_1
#> 17 B Group_1
#> 18 B Group_1
#> 19 B Group_2
#> 20 B Group_2
#> 21 B Group_2
#> 22 B Group_2
#> 23 B Group_2
#> 24 B Group_2
#> 25 B Group_2
#> 26 C Group_1
#> 27 C Group_1
#> 28 C Group_1
#> 29 C Group_1
#> 30 C Group_1
#> 31 C Group_1
#> 32 C Group_1
#> 33 C Group_1
#> 34 C Group_2
#> 35 C Group_2
#> 36 C Group_2
#> 37 C Group_2
#> 38 C Group_2
#> 39 C Group_2
#> 40 C Group_2
#> 41 C Group_2
#> 42 C Group_2
# modifying GreyMerchant's solution
library(magrittr)
my_datas2 %>%
tidyr::gather(key = "key",
value = "value",
-Answers) %$%
data.frame(Answers = rep(Answers, value),
Group = rep(key, value))
#> Answers Group
#> 1 A Group_1
#> 2 A Group_1
#> 3 A Group_1
#> 4 A Group_1
#> 5 A Group_1
#> 6 A Group_1
#> 7 A Group_1
#> 8 A Group_1
#> 9 B Group_1
#> 10 B Group_1
#> 11 B Group_1
#> 12 B Group_1
#> 13 B Group_1
#> 14 B Group_1
#> 15 C Group_1
#> 16 C Group_1
#> 17 C Group_1
#> 18 C Group_1
#> 19 C Group_1
#> 20 C Group_1
#> 21 C Group_1
#> 22 C Group_1
#> 23 A Group_2
#> 24 A Group_2
#> 25 A Group_2
#> 26 A Group_2
#> 27 B Group_2
#> 28 B Group_2
#> 29 B Group_2
#> 30 B Group_2
#> 31 B Group_2
#> 32 B Group_2
#> 33 B Group_2
#> 34 C Group_2
#> 35 C Group_2
#> 36 C Group_2
#> 37 C Group_2
#> 38 C Group_2
#> 39 C Group_2
#> 40 C Group_2
#> 41 C Group_2
#> 42 C Group_2
Thank you @Yarnabrina, I think everything is all right as I asked here before:
and you provided the solution to it. Additionally you provided another solutions which I am grateful for because I have learnt a lot today.
best regards,
A
Glad this worked for you ![]() So
So do.call is a really helpful function. You're able to provide it with a function and then you can give it a list and essentially it will repeat that function for the set and return it. You can see in this simple example below that combined2 does the rbind calls once for all three sets that were in a list instead of combining them separate.
one <- sample(c(1:10),10)
two <- sample(c(20:30),10)
three <- sample(c(40:50),10)
combined1 <- rbind(one,two,three)
combined1
#> [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
#> one 6 2 4 9 10 1 8 3 7 5
#> two 27 23 28 20 21 22 24 29 26 30
#> three 40 42 49 41 44 43 47 45 46 50
combination <- list(one, two, three)
combined2 <- do.call(rbind,combination)
combined2
#> [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
#> [1,] 6 2 4 9 10 1 8 3 7 5
#> [2,] 27 23 28 20 21 22 24 29 26 30
#> [3,] 40 42 49 41 44 43 47 45 46 50
Created on 2021-11-07 by the reprex package (v2.0.0)
mapply(rep,dat$answers,dat$value) Is slightly different. It is taking a function rep and then doing that function for each pair of values found within the two vectors. So it is taking answers and then seeing how many times it needs to repeat it for value and then this process repeats for the next pair of values found in answers and value
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.