Hi ALL, I need please help with this:
I have this data and code:
data_data_long <- structure(list(occurrence = c("yes", "yes", "yes", "no", "no",
"no", "yes", "yes", "yes", "no", "no", "no", "yes", "yes", "yes",
"no", "no", "no", "yes", "yes", "yes", "no", "no", "no", "yes",
"yes", "yes", "no", "no", "no", "yes", "yes", "yes", "no", "no",
"no", "yes", "yes", "yes", "no", "no", "no", "yes", "yes", "yes",
"no", "no", "no"), group = structure(c(1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), levels = c("Ia", "IIa"
), class = "factor"), sex = structure(c(1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), levels = c("Female",
"Male"), class = "factor"), side = structure(c(1L, 1L, 1L, 1L,
1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L), levels = c("left",
"right"), class = "factor"), n = c(128, 128, 128, 128, 128, 128,
128, 128, 128, 128, 128, 128, 84, 84, 84, 84, 84, 84, 84, 84,
84, 84, 84, 84, 131, 131, 131, 131, 131, 131, 131, 131, 131,
131, 131, 131, 71, 71, 71, 71, 71, 71, 71, 71, 71, 71, 71, 71
), symptom = structure(c(1L, 2L, 3L, 1L, 2L, 3L, 1L, 2L, 3L,
1L, 2L, 3L, 1L, 2L, 3L, 1L, 2L, 3L, 1L, 2L, 3L, 1L, 2L, 3L, 1L,
2L, 3L, 1L, 2L, 3L, 1L, 2L, 3L, 1L, 2L, 3L, 1L, 2L, 3L, 1L, 2L,
3L, 1L, 2L, 3L, 1L, 2L, 3L), levels = c("r1", "r2", "r3"), class = "factor"),
frequency = c(47, 32, 57, 81, 96, 71, 49, 27, 68, 79, 101,
60, 44, 35, 56, 40, 49, 28, 45, 35, 55, 39, 49, 29, 44, 30,
58, 87, 101, 73, 47, 34, 72, 84, 97, 59, 30, 29, 41, 41,
42, 30, 36, 30, 40, 35, 41, 31)), row.names = c(NA, -48L), class = c("tbl_df",
"tbl", "data.frame"))
symptoms <- data_data_long %>% select(symptom) %>% distinct() %>% unlist()
sex <- data_data_long %>% select(sex) %>% distinct() %>% unlist()
side <- data_data_long %>% select(side) %>% distinct() %>% unlist()
names_of_columns <- c("Symptoms","Sex","Side","Group","Incidence","n","Incidence_risk", "Incidence_risk_CIll", "Incidence_risk_CIul", "Incidence_OR",
"Incidence_OR_CIll", "Incidence_OR_CIul",
"x2", "pvalue")
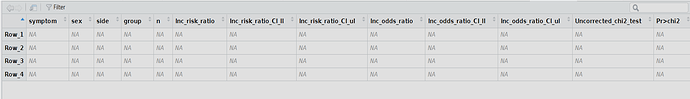
my_matrix <- matrix(NA, nrow = length( symptoms) * length(sex) * length(side) ,
ncol = length(names_of_columns), dimnames= list(c(), names_of_columns ))
rownames(my_matrix) <- paste0("Row_", seq_len(nrow(my_matrix)))
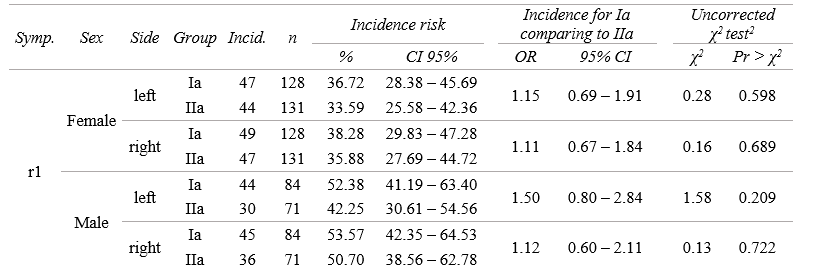
I want to fill in this earlier prepared empty matrix using for loop in R so I want to get results in my_matrix with Symptoms from r1 to r3, Sex: Female and Male, Side: Left and Right, Group: Ia or IIa, Incidence that is incidence risk, n – that is the number of participants in group, Incidence_risk that is incidence risk, Incidence_risk_CIll – that is confidence intervals for Incidence risk lower limit, Incidence_risk_CIul - that is confidence intervals for Incidence risk upper limit, Incidence_OR- that is inc odds ratio, and subsequently confidence intervals for Incidence odds ratio lower and upper limit, x2 which is value of chi square statistics and pvalue of chi square test.
All these results must be extracted from output of this function:
library(epiR)
data_data_long %>% dplyr::filter( sex == "Female", side =="left", symptom =="r1") %>%
dplyr::select(frequency) %>% unlist() %>%
matrix(., ncol =2, dimnames = list(c("Occurrence", "Not occurrence"), c("Ia", "IIa"))) %>% t() %>%
epi.2by2(dat = ., method = "cohort.count", conf.level = 0.95, units = 100,
interpret = FALSE, outcome = "as.columns")
and so on for r2 and r3, for female and male, group Ia and IIa, side: left and right, etc. Please help. Or maybe you know a way without using for loop ? Thank you.