Hi All,
I have got a dataframe dfSamp2. I wanted to calculate chisq.test() when grouping variable is set to "week".
So far I have done a for loop, which has given me this:
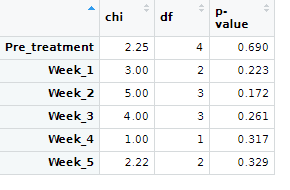
I am OK with it, below please find a workind code.
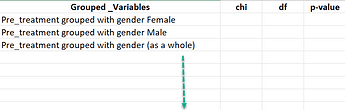
My question is how do I modify this for loop below, to be grouped by two variables like: treatment and gender, please ?
My desired result would be something like this with all possible combinations of grouping variables I hope.:
All help will be greatly appreciated, thank you.
library(tidyverse)
dfSamp2 <- structure(list(
center_id = c(
"50777", "07057", "50777", "50777",
"14659", "51238", "43437", "51238", "51238", "43437", "35702",
"50777", "50777", "50097", "51238", "43437", "14659", "50777",
"50777", "50777", "14659", "14659", "14659", "51238", "43437",
"50777", "14659", "35702", "43437", "35702"
), center_size = structure(c(
3L,
1L, 3L, 3L, 2L, 2L, 3L, 2L, 2L, 3L, 2L, 3L, 3L, 3L, 2L, 3L, 2L,
3L, 3L, 3L, 2L, 2L, 2L, 2L, 3L, 3L, 2L, 2L, 3L, 2L
), .Label = c(
"Small",
"Medium", "Large"
), class = "factor"), gender = structure(c(
2L,
1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 1L, 2L,
1L, 2L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 2L, 1L
), .Label = c(
"Male",
"Female"
), class = "factor"), treatment = structure(c(
1L, 2L,
2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 1L,
1L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 1L, 2L, 1L
), .Label = c(
"Anticonvulsant",
"Placebo"
), class = "factor"), week = structure(c(
2L, 2L, 3L,
4L, 6L, 5L, 4L, 5L, 6L, 1L, 3L, 6L, 1L, 4L, 1L, 3L, 4L, 6L, 6L,
1L, 5L, 1L, 5L, 3L, 3L, 1L, 2L, 1L, 1L, 1L
), .Label = c(
"Pre_treatment",
"Week_1", "Week_2", "Week_3", "Week_4", "Week_5"
), class = "factor"),
convulsions = c(
10, 4, 2, 2, 13, 2, 5, 2, 2, 4, 1, 2, 2,
4, 3, 1, 0, 2, 1, 27, 7, 1, 8, 2, 24, 2, 7, 10, 12, 13
)
), variable.labels = c(
center_id = "Center ID",
center_size = "Center size", gender = "Gender",
treatment = "Treatment received", week = "Week", convulsions = "Number of convulsions"
), codepage = 65001L, row.names = c(
198L,
115L, 295L, 416L, 580L, 525L, 365L, 540L, 656L, 43L, 243L, 641L,
79L, 391L, 90L, 263L, 349L, 629L, 628L, 64L, 464L, 15L, 466L,
321L, 258L, 83L, 134L, 23L, 47L, 28L
), class = "data.frame")
col_vars <- c("chi", "df", "p-value")
row_vars <- c(unique(dfSamp2$week)) %>% as.vector() %>% sort()
mydf <- matrix(NA, nrow = length(unique(dfSamp2$week)), ncol = length(col_vars), dimnames = list(row_vars, col_vars))
for(i in seq(row_vars)){
df_temp <- dfSamp2 %>% select(center_id, gender, week) %>% filter(week == row_vars[i])
# assingning chi
mydf[i,1] <- round(chisq.test(df_temp$center_id, df_temp$gender)$statistic, 2)
#assingning df
mydf[i,2] <-chisq.test(df_temp$center_id, df_temp$gender)$parameter
# assingning p-value
temp <- round(chisq.test(df_temp$center_id, df_temp$gender)$p.value, 3)
if (temp < 0.001) temp <- "< 0.001"
mydf[i,3] <- temp
}
Created on 2022-02-21 by the reprex package (v2.0.1)