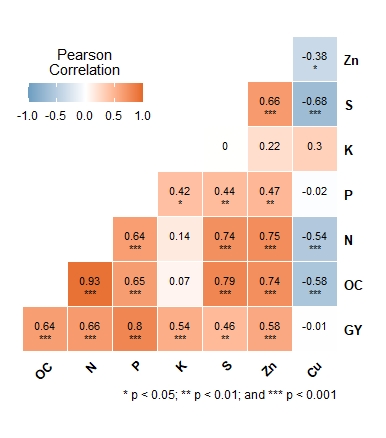
Hi, I've tried to create a correlation matrix plot using the following data and codes.
Treat
Year
Rep
GY
OC
N
P
K
S
Zn
Cu
Control
2007
1
1.82
8.20
0.90
5.00
0.09
4.50
2.00
4.60
Control
2007
2
1.84
10.50
0.70
6.00
0.07
6.00
1.30
3.60
Control
2007
3
1.79
9.20
1.10
4.00
0.11
3.30
1.50
5.00
Rev. control
2007
1
10.57
12.30
1.20
11.00
0.13
5.10
5.50
5.60
Rev. control
2007
2
10.60
14.20
1.40
14.00
0.15
6.00
6.30
3.90
Rev. control
2007
3
10.63
10.40
1.00
17.00
0.11
4.50
4.10
7.00
NPK
2007
1
9.92
11.60
1.10
24.00
0.10
6.10
2.60
5.50
NPK
2007
2
9.93
10.00
0.90
21.00
0.15
7.10
1.80
6.00
NPK
2007
3
9.93
10.80
1.30
24.00
0.19
5.10
3.40
4.10
NPKS
2007
1
10.56
13.00
1.30
14.00
0.10
7.00
5.20
5.60
NPKS
2007
2
10.62
13.60
1.40
12.00
0.14
4.10
6.50
6.00
NPKS
2007
3
10.44
12.70
1.20
16.00
0.12
5.10
3.90
4.90
NPKSZn
2007
1
10.77
16.00
1.50
28.00
0.11
9.00
3.60
5.60
NPKSZn
2007
2
10.73
15.00
1.40
30.00
0.14
11.00
2.50
6.00
NPKSZn
2007
3
10.81
14.00
1.60
26.00
0.11
7.00
4.70
5.50
NPKSZnCu
2007
1
11.56
14.00
1.20
20.00
0.16
8.50
4.00
9.60
NPKSZnCu
2007
2
11.46
13.20
1.40
22.00
0.14
10.00
3.00
7.60
NPKSZnCu
2007
3
11.65
14.50
1.60
24.00
0.12
7.00
4.70
11.00
Control
2017
1
3.36
12.87
1.05
5.81
0.08
10.13
3.06
0.58
Control
2017
2
3.53
12.48
1.33
7.61
0.08
10.44
2.91
0.60
Control
2017
3
3.45
12.87
1.05
8.08
0.08
11.45
2.70
0.60
Rev. control
2017
1
11.99
14.43
1.61
17.20
0.13
14.45
4.18
1.15
Rev. control
2017
2
11.33
14.82
1.68
15.95
0.14
16.15
4.31
1.13
Rev. control
2017
3
12.14
15.60
1.54
15.00
0.10
19.41
4.25
1.20
NPK
2017
1
11.91
18.72
1.96
23.45
0.09
10.21
4.01
0.53
NPK
2017
2
11.12
19.11
1.82
23.36
0.13
10.20
4.43
0.53
NPK
2017
3
11.74
19.50
2.03
23.72
0.09
11.44
4.27
0.53
NPKS
2017
1
11.48
20.67
2.10
23.90
0.10
15.26
6.22
0.55
NPKS
2017
2
11.16
20.67
2.03
22.40
0.13
14.91
5.94
0.55
NPKS
2017
3
12.05
20.67
2.38
24.50
0.13
14.37
6.26
0.55
NPKSZn
2017
1
12.42
20.28
1.82
29.72
0.13
19.52
10.90
0.60
NPKSZn
2017
2
12.06
19.89
2.24
23.86
0.12
16.31
9.88
0.58
NPKSZn
2017
3
12.62
20.28
2.10
20.44
0.13
17.63
11.13
0.55
NPKSZnCu
2017
1
12.61
19.50
2.17
25.61
0.12
17.52
11.74
1.43
NPKSZnCu
2017
2
12.44
19.11
2.38
27.20
0.12
15.89
11.15
1.40
NPKSZnCu
2017
3
12.39
20.67
2.03
25.99
0.14
17.93
11.76
1.48
library(readxl)#6D9EC1 ", mid = "white", high ="#E46726 ", midpoint = 0,
And I've got the below plot.
what R2 values ?
Hi Nirgrahamuk,
I think you may have confused the matter by attachingR which presumably has a 'values' field ? otherwise its not clear how +goem_text is getting values.
but we can see that the plot.corr_coef has a digits.cor option, here I do it twice once with 1 digit and once with 3
library(metan)
cc <- corr_coef(data_ge2[,1:12])
plot(cc,reorder = FALSE,
digits.cor = 1)
plot(cc,reorder = FALSE,
digits.cor = 3)
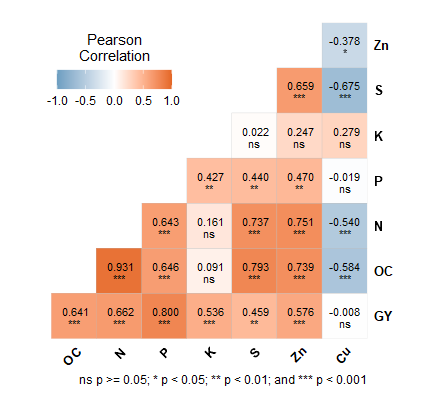
Sorry for the geom_text, it was a mistake. digits.cor worked but still the problem is not solved when the last decimal places are zero e.g. 0.30 is showing as 0.3 and 0.00 is showing as 0. Is there any solution?
I cant reproduce that behaviour.
Hi, here is the reprex.
library(readxl)
library(metan)
P<-plot(CM,reorder = FALSE, digits.cor = 3)+#6D9EC1 ", mid = "white", high ="#E46726 ", midpoint = 0,
P
I had to alter what you provided so as to be runnable.
library(tidyverse)
library(metan)
R <- readr::read_delim("Treat Year Rep GY OC N P K S Zn Cu
Control 2007 1 1.82 8.20 0.90 5.00 0.09 4.50 2.00 4.60
Control 2007 2 1.84 10.50 0.70 6.00 0.07 6.00 1.30 3.60
Control 2007 3 1.79 9.20 1.10 4.00 0.11 3.30 1.50 5.00
Rev.control 2007 1 10.57 12.30 1.20 11.00 0.13 5.10 5.50 5.60
Rev.control 2007 2 10.60 14.20 1.40 14.00 0.15 6.00 6.30 3.90
Rev.control 2007 3 10.63 10.40 1.00 17.00 0.11 4.50 4.10 7.00
NPK 2007 1 9.92 11.60 1.10 24.00 0.10 6.10 2.60 5.50
NPK 2007 2 9.93 10.00 0.90 21.00 0.15 7.10 1.80 6.00
NPK 2007 3 9.93 10.80 1.30 24.00 0.19 5.10 3.40 4.10
NPKS 2007 1 10.56 13.00 1.30 14.00 0.10 7.00 5.20 5.60
NPKS 2007 2 10.62 13.60 1.40 12.00 0.14 4.10 6.50 6.00
NPKS 2007 3 10.44 12.70 1.20 16.00 0.12 5.10 3.90 4.90
NPKSZn 2007 1 10.77 16.00 1.50 28.00 0.11 9.00 3.60 5.60
NPKSZn 2007 2 10.73 15.00 1.40 30.00 0.14 11.00 2.50 6.00
NPKSZn 2007 3 10.81 14.00 1.60 26.00 0.11 7.00 4.70 5.50
NPKSZnCu 2007 1 11.56 14.00 1.20 20.00 0.16 8.50 4.00 9.60
NPKSZnCu 2007 2 11.46 13.20 1.40 22.00 0.14 10.00 3.00 7.60
NPKSZnCu 2007 3 11.65 14.50 1.60 24.00 0.12 7.00 4.70 11.00
Control 2017 1 3.36 12.87 1.05 5.81 0.08 10.13 3.06 0.58
Control 2017 2 3.53 12.48 1.33 7.61 0.08 10.44 2.91 0.60
Control 2017 3 3.45 12.87 1.05 8.08 0.08 11.45 2.70 0.60
Rev.control 2017 1 11.99 14.43 1.61 17.20 0.13 14.45 4.18 1.15
Rev.control 2017 2 11.33 14.82 1.68 15.95 0.14 16.15 4.31 1.13
Rev.control 2017 3 12.14 15.60 1.54 15.00 0.10 19.41 4.25 1.20
NPK 2017 1 11.91 18.72 1.96 23.45 0.09 10.21 4.01 0.53
NPK 2017 2 11.12 19.11 1.82 23.36 0.13 10.20 4.43 0.53
NPK 2017 3 11.74 19.50 2.03 23.72 0.09 11.44 4.27 0.53
NPKS 2017 1 11.48 20.67 2.10 23.90 0.10 15.26 6.22 0.55
NPKS 2017 2 11.16 20.67 2.03 22.40 0.13 14.91 5.94 0.55
NPKS 2017 3 12.05 20.67 2.38 24.50 0.13 14.37 6.26 0.55
NPKSZn 2017 1 12.42 20.28 1.82 29.72 0.13 19.52 10.90 0.60
NPKSZn 2017 2 12.06 19.89 2.24 23.86 0.12 16.31 9.88 0.58
NPKSZn 2017 3 12.62 20.28 2.10 20.44 0.13 17.63 11.13 0.55
NPKSZnCu 2017 1 12.61 19.50 2.17 25.61 0.12 17.52 11.74 1.43
NPKSZnCu 2017 2 12.44 19.11 2.38 27.20 0.12 15.89 11.15 1.40
NPKSZnCu 2017 3 12.39 20.67 2.03 25.99 0.14 17.93 11.76 1.48",delim=" ")
CM<-corr_coef(R[4:11])
CM
P<-plot(CM,reorder = FALSE, digits.cor = 3)+
theme(axis.text.x = element_text(color = "black",size = 10, face = "bold"))+
theme(axis.text.y = element_text(color = "black",size = 10, face = "bold"))+
theme(plot.margin = margin(0.6,0.6,0.6,0.6, "cm"))+
scale_fill_gradient2(low = "#6D9EC1", mid = "white", high ="#E46726", midpoint = 0,
limits = c(-1,1), space = "Lab",
name="Pearson\nCorrelation")
P
this produced
Thank you very much for your quick response. I really appreciate your feed back. I finally figure out the problem, it was with the metan package which I updated and everithing work perfect. Thanks again.
system
October 10, 2023, 7:57am
10
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.