The following library's are necessary
library(knitr)
library(rmarkdown)
library(kknn)
library(recipeselectors)
library(mice)
library(stats)
library(githubinstall)
library(xlsx)
library(tidymodels)
tidymodels_prefer(quiet = TRUE)
library(tidyverse)
library(conflicted)
#Set working directory
setwd("D:\PayStr\FindEngine\TidyModels\Data")
#Read in actual data set
org_data1 <- read.xlsx("Life Expectancy Data.xlsx", sheetIndex=1, header=TRUE, as.data.frame=TRUE)
#Dropping Country for now
org_data0<-org_data1%>%select(-Country)
org_data<-org_data0%>%filter(Y!=" ")
#Separating data into Training and Testing
set.seed(222)
Put 3/4 of the data into the training set
data_split <- initial_split(org_data, prop = 3/4)
Create data frames for the two sets:
train_data <- training(data_split)
test_data <- testing(data_split)
#Start our Recipe
LifeExp_rec <-
recipe(Y ~ ., data = train_data) %>%
step_dummy(all_nominal_predictors()) %>%
step_zv(all_predictors()) %>%
step_center(all_predictors()) %>%
step_scale(all_predictors()) %>%
step_impute_knn(all_predictors(), neighbors = 3) %>%
step_select_vip(all_predictors(), outcome = "Y", model = linear_reg, top_p = 12, threshold = 0.7)
#prep()
LifeExp_rec
#Specific Model
linear_model<-
linear_reg() %>%
set_engine("lm") %>%
set_mode("regression")
#parsnip::translate()
linear_model
#Set up Work Flow with Recipe and model
LifeExp_wflow<-
workflow() %>%
add_recipe(LifeExp_rec) %>%
add_model(linear_model)
LifeExp_wflow
#Run work flow to Fit training data
LifeExp_fit<-
LifeExp_wflow %>%
fit(data=train_data)
LifeExp_fit
#Extract parameters from Fit
LifeExp_fit %>%
extract_fit_parsnip() %>%
tidy()
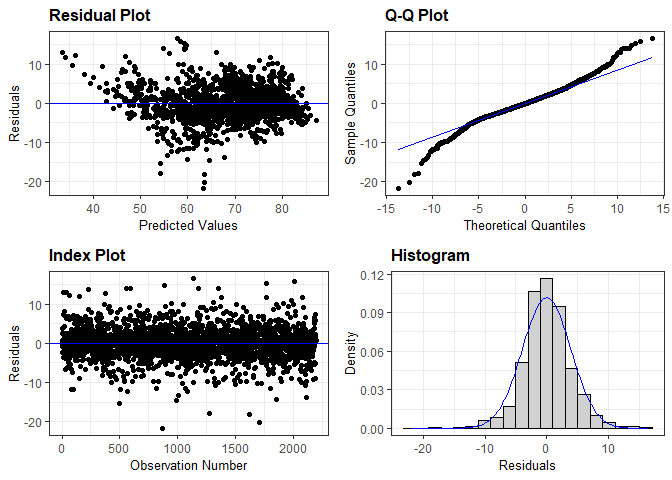
#Residual analysis
Life_Exp_aug<-
augment(LifeExp_fit, train_data)
Life_Exp_aug$.resid=(Life_Exp_aug$Y-Life_Exp_aug$.pred)
resid_auxpanel(Life_Exp_aug$.resid, Life_Exp_aug$.pred, bins = 20)
#Measures of model fit on Training data
Life_Exp_TR_P<-predict(LifeExp_fit,new_data = train_data)
Life_Exp_RMSE_TR<-bind_cols(Life_Exp_TR_P, train_data %>% select(Y))
Life_Exp_Metrics <-metric_set(rmse, rsq, mae)
Life_Exp_Metrics(Life_Exp_RMSE_TR, truth=Y, estimate=.pred)
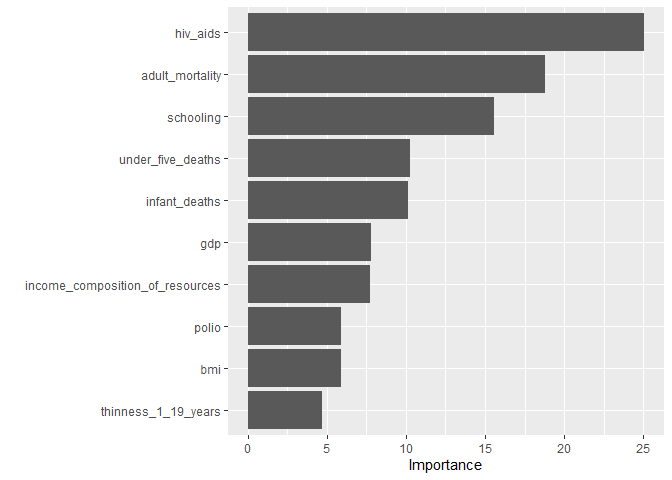
#Variable Importance
LifeExp_fit %>%
extract_fit_parsnip() %>%
vip(method = "model",
target = "Y", metric = "rmse",
pred_wrapper = stats::predict, train = train_data)
#Measures of model fit on Test data
Life_Exp_TT_P<-predict(LifeExp_fit, new_data = test_data)
Life_Exp_RMSE_TT<-bind_cols(Life_Exp_TT_P, test_data %>% select(Y))
Life_Exp_Metrics <-metric_set(rmse, rsq, mae)
Life_Exp_Metrics(Life_Exp_RMSE_TT, truth=Y, estimate=.pred)
#K-fold Cross validation
set.seed(1001)
Life_Exp_folds<- vfold_cv(train_data, v=10)
Life_Exp_folds
#Setting some resampling controls
keep_pred <-control_resamples(save_pred=TRUE, save_workflow= TRUE)
set.seed(1003)
#Fitting the resampled data
Life_Exp_Res<-
LifeExp_wflow %>%
fit_resamples(resamples=Life_Exp_folds, control=keep_pred)
Life_Exp_Res
#Collecting Metrics
collect_metrics(Life_Exp_Res)
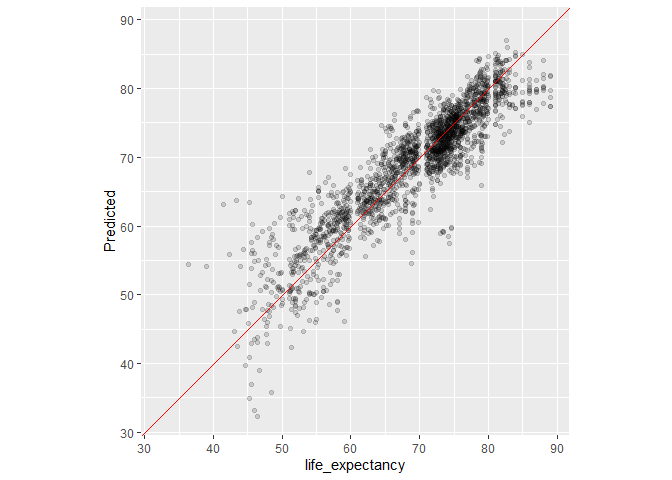
#Output the assessment predictions for plotting
assess_res<-collect_predictions(Life_Exp_Res)
assess_res
assess_res %>%
ggplot(aes(x = Y, y = .pred)) +
geom_point(alpha = .15) +
geom_abline(color = "red") +
coord_obs_pred() +
ylab("Predicted")