I am using the caret package to tune a Random Forest (RF) model using ranger. Because in the ranger package I can't tune the numer of trees, I am using the caret package. The metric to find the optimal number of trees is R-Squared. The range of trees I am testing is from 500 to 3000 with step 500 (500, 1000, 1500,..., 3000).
The issue is that the R-squared is the same for every number of tree (see the attached image below): 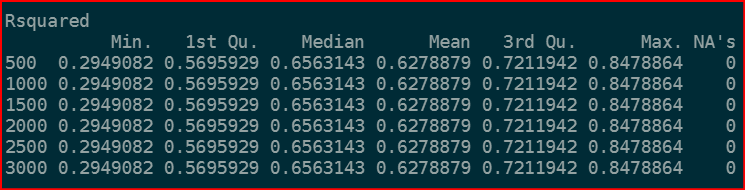
I don't think that's correct so I believe that there is something wrong with my code. Why am I getting the same R-squared?
Here is the code:
library(caret)
library(ranger)
# Load the data
block.data <- read.csv("path/block.data.csv")
eq1 = ntl ~ .
# Define the cross-validation method for hyperparameter tuning
control <- trainControl(method = "cv", number = 10, savePredictions = FALSE,
search = 'grid', allowParallel = TRUE)
# default model
rf_default = train(eq1,
data = block.data,
method = "ranger",
metric = "Rsquared",
trControl = control)
print(rf_default)
# Define the grid of hyperparameters to be tuned
tuneGrid <- expand.grid(mtry = c(2, 3, 4, 5, 6, 7), # number of predictor variables to sample at each split
splitrule = c("variance", "extratrees"), # splitting rule
min.node.size = c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10)) # minimum size of terminal nodes
# Train the model with hyperparameter tuning using caret
set.seed(234)
rf_model <- train(eq1, # formula for the response and predictors
data = block.data,
method = "ranger",
trControl = control,
tuneGrid = tuneGrid)
rf_model$bestTune
tuneGrid <- expand.grid(mtry = rf_model$bestTune$mtry,
splitrule = rf_model$bestTune$splitrule,
min.node.size = rf_model$bestTune$min.node.size)
store_maxtrees <- list()
for (ntree in c(500, 1000, 1500, 2000, 2500, 3000)) {
set.seed(345)
rf_maxtrees <- train(eq1,
data = block.data,
method = "ranger",
metric = "Rsquared",
tuneGrid = tuneGrid,
trControl = control,
ntree = ntree)
key <- toString(ntree)
store_maxtrees[[key]] <- rf_maxtrees
}
results_tree <- resamples(store_maxtrees)
summary(results_tree)