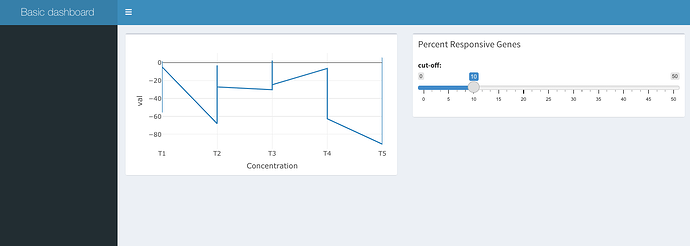
Hi,
I am looking to find the number of data points lying between a range of values by moving a slider in R shiny app. For instance, how many data points are in between 0-25, 0-50 etc. It seems like currently, the line plot is just plotting on the val column directly, however, I am interested to know the number of data points lying between the range of selected values on the slider. Is it possible to identify or a new filtered column must be added to a data in order to accomplish that?
Responsive Genes count in each concentration; For each concentration, responsive genes are set of genes that has been detected with a values between ranges.
Thank you,
Toufiq
dput(df)
structure(list(Samples = c("Gene_1", "Gene_2", "Gene_3", "Gene_4",
"Gene_5", "Gene_1", "Gene_2", "Gene_3", "Gene_4", "Gene_5", "Gene_1",
"Gene_2", "Gene_3", "Gene_4", "Gene_5", "Gene_1", "Gene_2", "Gene_3",
"Gene_4", "Gene_5", "Gene_1", "Gene_2", "Gene_3", "Gene_4", "Gene_5",
"Gene_1", "Gene_2", "Gene_3", "Gene_4", "Gene_5", "Gene_1", "Gene_2",
"Gene_3", "Gene_4", "Gene_5", "Gene_1", "Gene_2", "Gene_3", "Gene_4",
"Gene_5", "Gene_1", "Gene_2", "Gene_3", "Gene_4", "Gene_5"),
val = c(37.1, 75.3, 46, 96.33, 39, 58.22, 83, 11.36, 64,
-91, 38, -68, 28, 81, -53, 1.6, -3.3, -30.33, -6.65, 61.99,
94.01, 79.01, 2.2, 86, 14, 26, 93, 32, 76, 5.5, 69, 50.1,
98.2, 18.3, 74.4, -55.66, -33.33, -24.69, -62.69, -8.87,
-4.99, -27.22, 99.1, 41.01, -6.5), variable = c("category1",
"category1", "category1", "category1", "category1", "category2",
"category2", "category2", "category2", "category2", "category3",
"category3", "category3", "category3", "category3", "category4",
"category4", "category4", "category4", "category4", "category5",
"category5", "category5", "category5", "category5", "category6",
"category6", "category6", "category6", "category6", "category7",
"category7", "category7", "category7", "category7", "category8",
"category8", "category8", "category8", "category8", "category9",
"category9", "category9", "category9", "category9"), Concentration = c("T1",
"T2", "T3", "T4", "T5", "T1", "T2", "T3", "T4", "T5", "T1",
"T2", "T3", "T4", "T5", "T1", "T2", "T3", "T4", "T5", "T1",
"T2", "T3", "T4", "T5", "T1", "T2", "T3", "T4", "T5", "T1",
"T2", "T3", "T4", "T5", "T1", "T2", "T3", "T4", "T5", "T1",
"T2", "T3", "T4", "T5")), class = "data.frame", row.names = c(NA,
-45L))
#> Samples val variable Concentration
#> 1 Gene_1 37.10 category1 T1
#> 2 Gene_2 75.30 category1 T2
#> 3 Gene_3 46.00 category1 T3
#> 4 Gene_4 96.33 category1 T4
#> 5 Gene_5 39.00 category1 T5
#> 6 Gene_1 58.22 category2 T1
#> 7 Gene_2 83.00 category2 T2
#> 8 Gene_3 11.36 category2 T3
#> 9 Gene_4 64.00 category2 T4
#> 10 Gene_5 -91.00 category2 T5
#> 11 Gene_1 38.00 category3 T1
#> 12 Gene_2 -68.00 category3 T2
#> 13 Gene_3 28.00 category3 T3
#> 14 Gene_4 81.00 category3 T4
#> 15 Gene_5 -53.00 category3 T5
#> 16 Gene_1 1.60 category4 T1
#> 17 Gene_2 -3.30 category4 T2
#> 18 Gene_3 -30.33 category4 T3
#> 19 Gene_4 -6.65 category4 T4
#> 20 Gene_5 61.99 category4 T5
#> 21 Gene_1 94.01 category5 T1
#> 22 Gene_2 79.01 category5 T2
#> 23 Gene_3 2.20 category5 T3
#> 24 Gene_4 86.00 category5 T4
#> 25 Gene_5 14.00 category5 T5
#> 26 Gene_1 26.00 category6 T1
#> 27 Gene_2 93.00 category6 T2
#> 28 Gene_3 32.00 category6 T3
#> 29 Gene_4 76.00 category6 T4
#> 30 Gene_5 5.50 category6 T5
#> 31 Gene_1 69.00 category7 T1
#> 32 Gene_2 50.10 category7 T2
#> 33 Gene_3 98.20 category7 T3
#> 34 Gene_4 18.30 category7 T4
#> 35 Gene_5 74.40 category7 T5
#> 36 Gene_1 -55.66 category8 T1
#> 37 Gene_2 -33.33 category8 T2
#> 38 Gene_3 -24.69 category8 T3
#> 39 Gene_4 -62.69 category8 T4
#> 40 Gene_5 -8.87 category8 T5
#> 41 Gene_1 -4.99 category9 T1
#> 42 Gene_2 -27.22 category9 T2
#> 43 Gene_3 99.10 category9 T3
#> 44 Gene_4 41.01 category9 T4
#> 45 Gene_5 -6.50 category9 T5
library(shiny)
library(shinydashboard)
library(plotly)
ui <- dashboardPage(
dashboardHeader(title = "Basic dashboard"),
dashboardSidebar(),
dashboardBody(
# Boxes need to be put in a row (or column)
fluidRow(
box(plotlyOutput("plotdf", height = 250)),
box(
title = "Responsive Genes Counts",
sliderInput(inputId = "val", label = "cut-off:", min = 0, max = 50, value = 10
)
)
)
)
)
server <- function(input, output) {
output$plotdf<-renderPlotly({
filter(df, val <= input$val) %>%
plot_ly() %>%
add_lines(type = 'scatter', mode = "lines",
x = ~Concentration, y = ~val)
})
}
shinyApp(ui, server)
Created on 2022-02-20 by the reprex package (v2.0.1)