Hi,
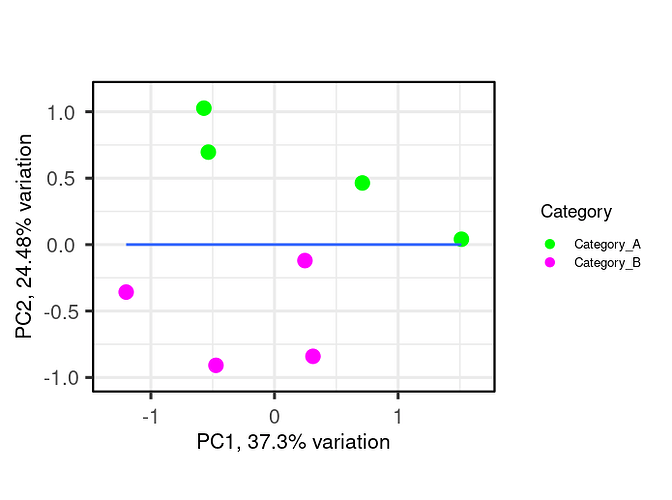
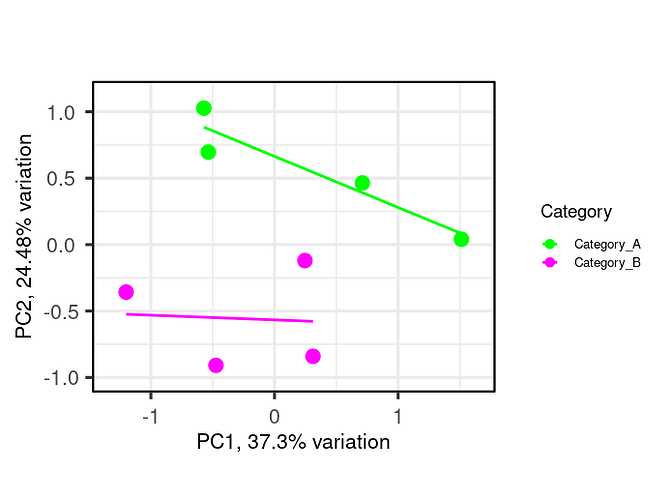
I am using R, I need some help on performing correlation analysis between 2 groups of samples (4 samples each) using a feature set of certain function. I want to create a scatter plot (XY plot) with regression line to visualize the correlation between the expression levels in Category_A vs. Category_B samples. I am working with the normalized intensity values, in order to study feature set of interest, I first subset my data by first mapping features of the feature set as rows (example below-Feature_set). Then, I plotted a pca plot using the below code (example-shown). Now I am interested to to draw regression line or a correlation over this pca plot.
Any other inputs and suggestions are also helpful.
Features as rows and samples as columns
(Feature_set)
Sample1 Sample2 Sample3
ABCC3 0.277954465 -0.02603594 0.19436366
ADAP1 0.068464085 -0.01052524 -0.17524178
AGR2 0.047788278 0.21902961 0.09946198
AKAP12 0.486130569 0.43430535 -1.24240970
ALDH3B2 -0.107000623 0.51769269 -0.49248772
ALOX5 -0.090541261 1.22612820 0.11739665
ANK2 -0.004727671 -0.04950311 -0.49840381
ANK3 0.493537146 0.36254862 0.29965816
ANXA4 -0.302597041 0.55604899 -0.15698546
ANXA9 -0.304823541 -0.14176106 -0.79107686
Sample4 Sample5 Sample6
ABCC3 -0.2376717 -0.01714223 -0.59277898
ADAP1 0.2441125 -0.31058118 0.10628536
AGR2 -0.1653739 0.82559878 0.54372150
AKAP12 -0.5335566 0.71488848 -0.96889009
ALDH3B2 -0.3321911 -0.06641259 0.03786791
ALOX5 -0.5616105 -0.51128082 -1.02459752
ANK2 -0.5445384 -0.71427818 -0.52205377
ANK3 0.8385503 -0.14598519 0.21803427
ANXA4 0.1398395 0.38658400 0.84453272
ANXA9 -0.3257577 -0.16644371 -0.16230293
Sample7 Sample8
ABCC3 -0.021885388 0.1928099
ADAP1 0.364137418 -0.2426156
AGR2 1.021608904 1.4078763
AKAP12 -0.427832989 -0.6140915
ALDH3B2 -0.604677137 0.6591462
ALOX5 0.506235945 -0.3515797
ANK2 -0.123123814 -0.8291402
ANK3 0.257584778 0.1990412
ANXA4 0.743738106 0.3672579
ANXA9 -0.005780198 -0.4071838
Sample metadata
Sample_metadata
#> Sample_ID Category
#> 1 Sample1 Category_A
#> 2 Sample2 Category_A
#> 3 Sample3 Category_A
#> 4 Sample4 Category_A
#> 5 Sample5 Category_B
#> 6 Sample6 Category_B
#> 7 Sample7 Category_B
#> 8 Sample8 Category_B
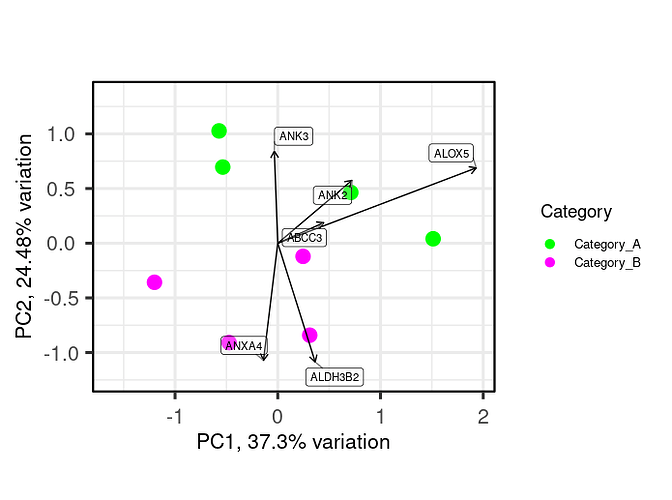
R code to plot pca
library(PCAtools)
p <- pca(Feature_set, metadata = Sample_metadata, removeVar = 0.1)
biplot(p,
lab = NULL,
x = 'PC1', y = 'PC2',
colby = 'Category', colkey = c('Category_A' = "green", "Category_B" = "magenta"),
legendPosition = 'right', legendLabSize = 10, legendIconSize = 3.0,
pointSize = 5)
Expected plot

Best Regards,
Toufiq