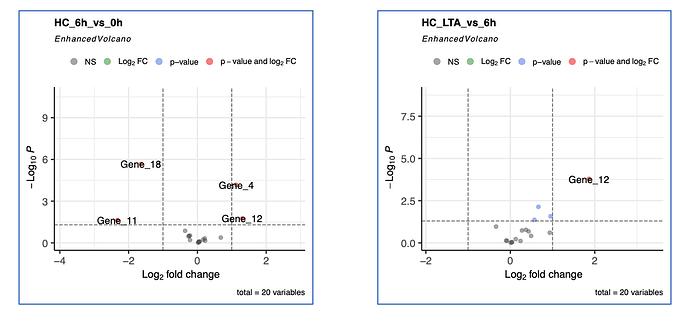
Hi,
I am working with matrix in R containing the quantitative data and trying to plot a volcano plot using library(EnhancedVolcano) package. I am currently analyzing by subsetting based on corresponding matching pairs of "Coef" and "P.value" (for instance; Coef.HC_6h_vs_0h and P.value.HC_6h_vs_0h) individually and export the pdf plot. I have large dataset containing many matching paired columns, this process becomes very tedious and cumbersome. I am exploring a way to loop to subset the data on each matching pairs of "Coef" and "P.value" generate separate plots in the form of the pdf file. I have provided the input data and expected results below. Please advise on how to solve this.
dput(Data)
structure(list(Coef.HC_6h_vs_0h = c(NA, NA, 0.048151066, 1.130642422,
0.68074318, 0.063779224, -0.358027426, -0.241326901, 0.167703878,
-0.257097748, -2.327977345, 1.300506434, 0.037118733, 0.036362435,
0.018953335, -0.215582086, -0.215130266, -1.642467234, 0.225615042,
0.212244164), Coef.HC_LTA_vs_6h = c(NA, NA, NA, 0.571320992,
0.240900855, 0.664981117, -0.080211466, 0.007521568, 0.032402696,
0.369452449, 0.934659157, 1.861006387, -0.338416494, 0.127921196,
0.040630241, 0.275547134, 0.952987298, 0.433537994, 0.492574777,
-0.099178916), Coef.UC_Rem_LPS_vs_6h = c(-0.259935478, -0.105746142,
0.207159012, 1.147068093, 0.211020926, 0.532359869, -0.101653129,
0.20209878, 0.016125514, 1.32593494, 1.496175233, 2.104156173,
-0.667087007, 0.154297917, 0.272351994, 0.791148254, 0.35998438,
1.399002196, 0.196009571, 0.619367312), Coef.CD_Mil_FLA_vs_6h = c(0.261472621,
-0.25263016, -0.041481308, -0.096265831, 0.004722895, 0.084040658,
-0.048078762, 0.009246748, -0.234490949, 0.273803791, 1.480274922,
0.292870886, -0.143628372, 0.087706716, -0.002301245, 0.15003941,
-0.314001872, 0.24746456, 0.617210833, -0.018220783), P.value.HC_6h_vs_0h = c(0.545376281,
0.011772197, 0.821228333, 6.67e-05, 0.415510885, 0.796781833,
0.136254462, 0.313415856, 0.589642204, 0.337351766, 0.023366465,
0.017807501, 0.860263888, 0.882854509, 0.956424822, 0.299125336,
0.622690737, 2.11e-06, 0.691578797, 0.474612129), P.value.HC_LTA_vs_6h = c(0.863203348,
0.465183842, 0.064113571, 0.043226481, 0.768757045, 0.00736856,
0.738392724, 0.97492814, 0.916995649, 0.168106123, 0.249987701,
0.000163064, 0.108745568, 0.598176335, 0.906755014, 0.184550004,
0.026536555, 0.208004113, 0.386500216, 0.73827056), P.value.UC_Rem_LPS_vs_6h = c(0.580841515,
0.728198309, 0.331085948, 5.23e-05, 0.796764557, 0.031810205,
0.672110689, 0.398510201, 0.958635746, 8.81e-07, 0.107068282,
1.5e-05, 0.001603959, 0.52498963, 0.432437774, 0.000146689, 0.410210239,
5.2e-05, 0.73035002, 0.037133718), P.value.CD_Mil_FLA_vs_6h = c(0.564348495,
0.406476956, 0.845654459, 0.733110608, 0.995400367, 0.73437091,
0.841329565, 0.969180147, 0.450794691, 0.306914258, 0.148066231,
0.558742168, 0.49581594, 0.717831127, 0.994706662, 0.469835936,
0.456921237, 0.47220793, 0.277934735, 0.951048936)), class = "data.frame", row.names = c("Gene_1",
"Gene_2", "Gene_3", "Gene_4", "Gene_5", "Gene_6", "Gene_7", "Gene_8",
"Gene_9", "Gene_10", "Gene_11", "Gene_12", "Gene_13", "Gene_14",
"Gene_15", "Gene_16", "Gene_17", "Gene_18", "Gene_19", "Gene_20"
))
#> Coef.HC_6h_vs_0h Coef.HC_LTA_vs_6h Coef.UC_Rem_LPS_vs_6h
#> Gene_1 NA NA -0.25993548
#> Gene_2 NA NA -0.10574614
#> Gene_3 0.04815107 NA 0.20715901
#> Gene_4 1.13064242 0.571320992 1.14706809
#> Gene_5 0.68074318 0.240900855 0.21102093
#> Gene_6 0.06377922 0.664981117 0.53235987
#> Gene_7 -0.35802743 -0.080211466 -0.10165313
#> Gene_8 -0.24132690 0.007521568 0.20209878
#> Gene_9 0.16770388 0.032402696 0.01612551
#> Gene_10 -0.25709775 0.369452449 1.32593494
#> Gene_11 -2.32797734 0.934659157 1.49617523
#> Gene_12 1.30050643 1.861006387 2.10415617
#> Gene_13 0.03711873 -0.338416494 -0.66708701
#> Gene_14 0.03636243 0.127921196 0.15429792
#> Gene_15 0.01895334 0.040630241 0.27235199
#> Gene_16 -0.21558209 0.275547134 0.79114825
#> Gene_17 -0.21513027 0.952987298 0.35998438
#> Gene_18 -1.64246723 0.433537994 1.39900220
#> Gene_19 0.22561504 0.492574777 0.19600957
#> Gene_20 0.21224416 -0.099178916 0.61936731
#> Coef.CD_Mil_FLA_vs_6h P.value.HC_6h_vs_0h P.value.HC_LTA_vs_6h
#> Gene_1 0.261472621 0.54537628 0.863203348
#> Gene_2 -0.252630160 0.01177220 0.465183842
#> Gene_3 -0.041481308 0.82122833 0.064113571
#> Gene_4 -0.096265831 0.00006670 0.043226481
#> Gene_5 0.004722895 0.41551088 0.768757045
#> Gene_6 0.084040658 0.79678183 0.007368560
#> Gene_7 -0.048078762 0.13625446 0.738392724
#> Gene_8 0.009246748 0.31341586 0.974928140
#> Gene_9 -0.234490949 0.58964220 0.916995649
#> Gene_10 0.273803791 0.33735177 0.168106123
#> Gene_11 1.480274922 0.02336646 0.249987701
#> Gene_12 0.292870886 0.01780750 0.000163064
#> Gene_13 -0.143628372 0.86026389 0.108745568
#> Gene_14 0.087706716 0.88285451 0.598176335
#> Gene_15 -0.002301245 0.95642482 0.906755014
#> Gene_16 0.150039410 0.29912534 0.184550004
#> Gene_17 -0.314001872 0.62269074 0.026536555
#> Gene_18 0.247464560 0.00000211 0.208004113
#> Gene_19 0.617210833 0.69157880 0.386500216
#> Gene_20 -0.018220783 0.47461213 0.738270560
#> P.value.UC_Rem_LPS_vs_6h P.value.CD_Mil_FLA_vs_6h
#> Gene_1 0.580841515 0.5643485
#> Gene_2 0.728198309 0.4064770
#> Gene_3 0.331085948 0.8456545
#> Gene_4 0.000052300 0.7331106
#> Gene_5 0.796764557 0.9954004
#> Gene_6 0.031810205 0.7343709
#> Gene_7 0.672110689 0.8413296
#> Gene_8 0.398510201 0.9691801
#> Gene_9 0.958635746 0.4507947
#> Gene_10 0.000000881 0.3069143
#> Gene_11 0.107068282 0.1480662
#> Gene_12 0.000015000 0.5587422
#> Gene_13 0.001603959 0.4958159
#> Gene_14 0.524989630 0.7178311
#> Gene_15 0.432437774 0.9947067
#> Gene_16 0.000146689 0.4698359
#> Gene_17 0.410210239 0.4569212
#> Gene_18 0.000052000 0.4722079
#> Gene_19 0.730350020 0.2779347
#> Gene_20 0.037133718 0.9510489
library(EnhancedVolcano)
## 'HC_6h_vs_0h'
pdf("Plot_for_HC_6h_vs_0h.pdf",height = 7, width = 7)
EnhancedVolcano(Data,
lab = rownames(Data),
x = 'Coef.HC_6h_vs_0h',
y = 'P.value.HC_6h_vs_0h',
title = 'HC_6h_vs_0h',
pCutoff = 0.05,
FCcutoff = 1.0,
pointSize = 3.0,
labSize = 6.0)
dev.off()
## 'HC_LTA_vs_6h'
pdf("Plot_for_HC_LTA_vs_6h.pdf",height = 7, width = 7)
EnhancedVolcano(Data,
lab = rownames(Data),
x = 'Coef.HC_LTA_vs_6h',
y = 'P.value.HC_LTA_vs_6h',
title = 'HC_LTA_vs_6h',
pCutoff = 0.05,
FCcutoff = 1.0,
pointSize = 3.0,
labSize = 6.0)
dev.off()
Created on 2022-04-21 by the reprex package (v2.0.1)
Thank you,
Toufiq