Hi!
I have two data frames, lets call them df1 and df2. Both data frames have missing values. If there is a value in df1, this value should be kept. If there is an NA in df1, the value should be filled with the one from df2. For further analysis, I joined the two data frames.
library(tidyverse)
df = data.frame("id" = c("A1", "A2", "A3", "A11", "A12", "A20", "A31", "A32", "A33"),
"X2004" = c(1, 0.5, 3, NA, 2, 1, 3, NA, 4),
"X2005" = c(1, 2, 2, NA, 2.5, 1, 0.5, NA, 4),
"X2006" = c(2, 1.5, NA, 2, 5, 1.5, NA, 2.5, 4),
"y_2005" = c(0.5, 2, NA, 3, 2.5, 1, 0.5, 2, NA),
"y_2006" = c(1, NA, 1.5, 2, NA, 1.5, 5, 2, 4))
The variables X2004, X2005 and X2006 are yearly data from df1, the variables y_2005 and y_2006 are from df2. Now I want to fill the missing data of X2005 by y_2005 and X2006 by y_2006.
In my real data, I would have a lot more years. Because of that, I tried to do a for loop, in which I create the variable names.
# As there are less years in df2, I make a list of all the years contained in df2.
years = df %>% select(starts_with("y_")) # get all the years of df2
names(years) = gsub("y_", "", names(years)) # delete the praefix of the variable name
years = colnames(years) # make a list of the column names
years = as.numeric(years) # define the years as numeric
# now I'm creating an empty data frame for the result of the data imputation
df_joined = data.frame("id" = c("A1", "A2", "A3", "A11", "A12", "A20", "A31", "A32", "A33"))
# that's the loop
for (year in years) {
value_df1 = paste0("X", year) # variable name from df1
value_df2 = paste0("y_", year) # variable name from df2
new_value = paste0("y", year) # define new variable name
df_ = df %>% mutate(new_value = ifelse(!is.na(value_df1), value_df1, value_df2))%>%
select("id", new_value)
df_joined = left_join(df_joined, df_, by = "id") # append the new values to the empty data frame
}
The problem is, that R doesn't understand, that value_df1 and value_df2 are meant to be the variable names, that's why the result is the following:
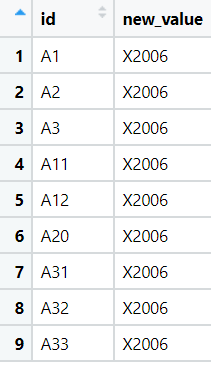
The mutate()-function itself works, when I execute the code with specific variables.
df_ = df %>% mutate(y2005= ifelse(!is.na(X2005), X2005, y_2005)) %>%
select("id", y2005)
I also tried it with the merge()-function, in that case, the variables are named the same:
# create df1 and df2
df1 = data.frame("id" = c("A1", "A2", "A3", "A11", "A12", "A20", "A31", "A32", "A33"),
"X2004" = c(1, 0.5, 3, NA, 2, 1, 3, NA, 4),
"X2005" = c(1, 2, 2, NA, 2.5, 1, 0.5, NA, 4),
"X2006" = c(2, 1.5, NA, 2, 5, 1.5, NA, 2.5, 4))
df2 = data.frame("id" = c("A1", "A2", "A3", "A11", "A12", "A20", "A31", "A32", "A33"),
"X2005" = c(0.5, 2, NA, 3, 2.5, 1, 0.5, 2, NA),
"X2006" = c(1, NA, 1.5, 2, NA, 1.5, 5, 2, 4))
# getting a list of the colnames
col_names = colnames(df2)
# merging df1 and df2
df = merge(df1, df2, by = c(col_names), all.x = TRUE)
If there are values in df1 and df2, R takes the values from df2. But I'd like to keep in these cases the values from df1 and only replace the NAs in df1 by the values from df2.
Do you have any ideas, how I could solve the problem?