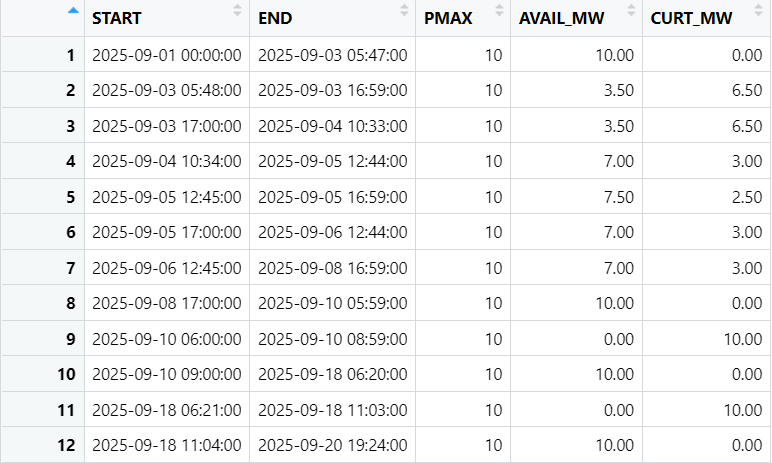
Hi everyone,
I'm having trouble having extracting minute intervals between two columns into a single column while having the same values of the other columns be the same within the interval.
So for the first row, it is transformed into
And those new rows will have PMAX = 10, AVAIL_MW = 10, CURT_MW = 0
library(data.table)
library(lubridate)
adj_av <- monthly_avail[, (OPR_TIME = seq(START, END, by = 'minute')), by= c("PMAX", "AVAIL_MW", "CURT_MW")]
Error in seq.POSIXt(START, END, by = "minute") :
Any help would be greatly appreciated!
FJCC
November 19, 2025, 4:54am
2
Here is an inelegant solution using base R functions. I used a smaller data set to save space.
DF <- data.frame(Start = as.POSIXct(c("2025-11-01 01:01:00", "2025-11-01 01:05:00", "2025-11-01 01:12:00")),
End = as.POSIXct(c("2025-11-01 01:04:00", "2025-11-01 01:11:00", "2025-11-01 01:14:00")),
PMAX = c(10,10,10), Avail_MW = c(10, 3.5,4), Curr_MW = c(0, 6.5,6))
DF
#> Start End PMAX Avail_MW Curr_MW
#> 1 2025-11-01 01:01:00 2025-11-01 01:04:00 10 10.0 0.0
#> 2 2025-11-01 01:05:00 2025-11-01 01:11:00 10 3.5 6.5
#> 3 2025-11-01 01:12:00 2025-11-01 01:14:00 10 4.0 6.0
Minutes <- (as.numeric(DF$End) - as.numeric(DF$Start))/60 + 1
StartVec <- seq.POSIXt(from = DF[1,1], to = DF[3,2], "min")
rep_date <- function(D,R) {
rep(D, R)
}
OutListEnd <- mapply(rep_date, DF$End, Minutes)
OutListEnd <- as.POSIXct(unlist(OutListEnd))
OutListPMax <- mapply(rep_date, DF$PMAX, Minutes)
OutListPMax <- unlist(OutListPMax)
OutListAvail_MW <- mapply(rep_date, DF$Avail_MW, Minutes)
OutListAvail_MW <- unlist(OutListAvail_MW)
OutListCurr_MW <- mapply(rep_date, DF$Curr_MW, Minutes)
OutListCurr_MW <- unlist(OutListCurr_MW)
data.frame(StartVec, OutListEnd,OutListPMax, OutListAvail_MW, OutListCurr_MW)
#> StartVec OutListEnd OutListPMax OutListAvail_MW
#> 1 2025-11-01 01:01:00 2025-11-01 01:04:00 10 10.0
#> 2 2025-11-01 01:02:00 2025-11-01 01:04:00 10 10.0
#> 3 2025-11-01 01:03:00 2025-11-01 01:04:00 10 10.0
#> 4 2025-11-01 01:04:00 2025-11-01 01:04:00 10 10.0
#> 5 2025-11-01 01:05:00 2025-11-01 01:11:00 10 3.5
#> 6 2025-11-01 01:06:00 2025-11-01 01:11:00 10 3.5
#> 7 2025-11-01 01:07:00 2025-11-01 01:11:00 10 3.5
#> 8 2025-11-01 01:08:00 2025-11-01 01:11:00 10 3.5
#> 9 2025-11-01 01:09:00 2025-11-01 01:11:00 10 3.5
#> 10 2025-11-01 01:10:00 2025-11-01 01:11:00 10 3.5
#> 11 2025-11-01 01:11:00 2025-11-01 01:11:00 10 3.5
#> 12 2025-11-01 01:12:00 2025-11-01 01:14:00 10 4.0
#> 13 2025-11-01 01:13:00 2025-11-01 01:14:00 10 4.0
#> 14 2025-11-01 01:14:00 2025-11-01 01:14:00 10 4.0
#> OutListCurr_MW
#> 1 0.0
#> 2 0.0
#> 3 0.0
#> 4 0.0
#> 5 6.5
#> 6 6.5
#> 7 6.5
#> 8 6.5
#> 9 6.5
#> 10 6.5
#> 11 6.5
#> 12 6.0
#> 13 6.0
#> 14 6.0
Created on 2025-11-18 with reprex v2.1.1
here's my dplyr/tidyr solution
library(dplyr)
library(tidyr)
df <- tribble(
~START, ~END, ~PMAX, ~AVAIL_MW, ~CURT_MW,
"2025-09-01 00:00:00", "2025-09-03 05:47:00", 10, 10.00, 0.00,
"2025-09-01 05:48:00", "2025-09-03 16:59:00", 10, 3.50, 6.50,
"2025-09-03 17:00:00", "2025-09-04 10:33_00", 10, 3.50, 6.50,
) |>
mutate(
START = as.POSIXct(START),
END= as.POSIXct(END))
df |>
rowwise() |>
mutate(TIME = list(seq(START, END, by = "min"))) |>
unnest(TIME) |>
select(TIME, PMAX, AVAIL_MW, CURT_MW)
Thank you everyone! It did exactly what I needed.
If anyone can figure how to make it work with data.table that would be appreciated! At this point, it's more of a curiosity about why it wasn't working.
data.table version:
dt[, .(TIME = seq( START, END, by = "min"), PMAX, AVAIL_MW, CURT_MW ), by = 1:nrow(dt)][, nrow := NULL][]