Fermentation Explorer: A Scientific App with Features for the Entire Shiny Community
Fermentation Explorer: A Scientific App with Features for the Entire Shiny Community
Authors: Timothy Hackmann
Abstract: Fermentation Explorer is an app used by microbiologists, but it has features useful to the entire Shiny community. Originally described in the journal Science Advances, the app allows microbiologists to explore microbes that carry out fermentation. For the Shiny community, it presents several new features for building apps, such as new types of interactive plots and UI elements.
Full Description:
Key Features
- Original version published in a scientific journal
- Interactive plots
- New UI elements for Shiny
- Navigation Buttons
- Large buttons with icon, title, and subtitle (ideal for a home page)
- Validation Modal
- Reports an error message if an input (e.g., uploaded file) is invalid
- Built on
shiny::validate()andshiny::showModal()
- File Input with Modal
- Built on
fileInput()and includes a link to a modal for more actions or information
- Built on
- Navigation Buttons
- Improved predictive tools for data science
- Metabolic networks and flux-balance analysis
- Built by extending base functions of
fbar
- Built by extending base functions of
- Predictions from taxonomy
- Built by implementing basic concept of FAPROTAX in R
- Predictions from machine learning
- Built using
randomForest
- Built using
- Metabolic networks and flux-balance analysis
- Video tutorials
Gallery
-
3D network graph
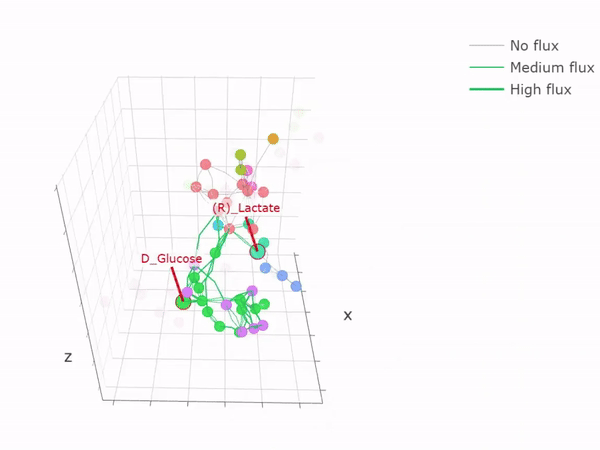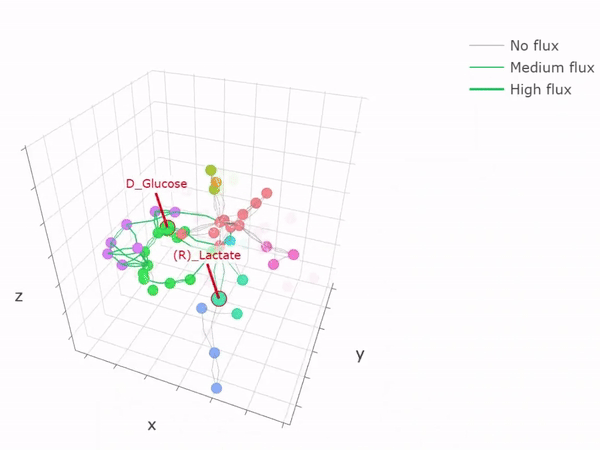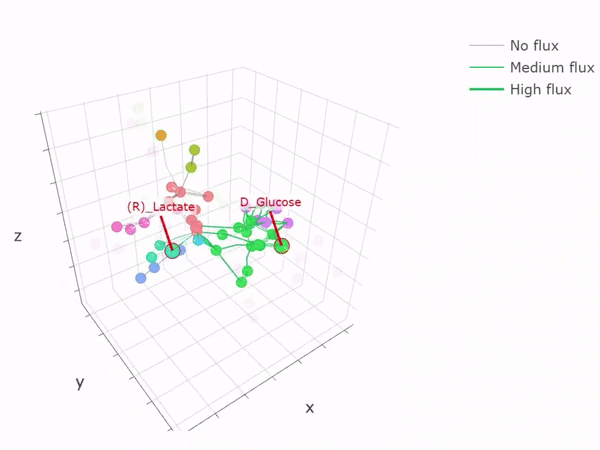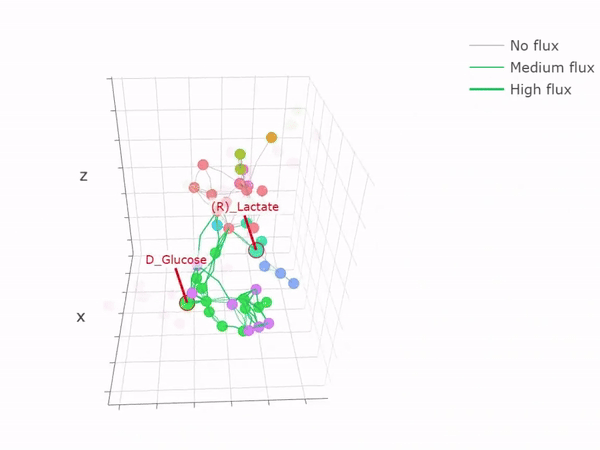
-
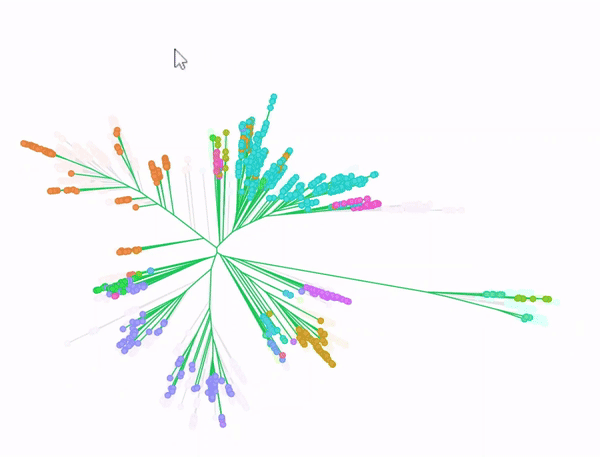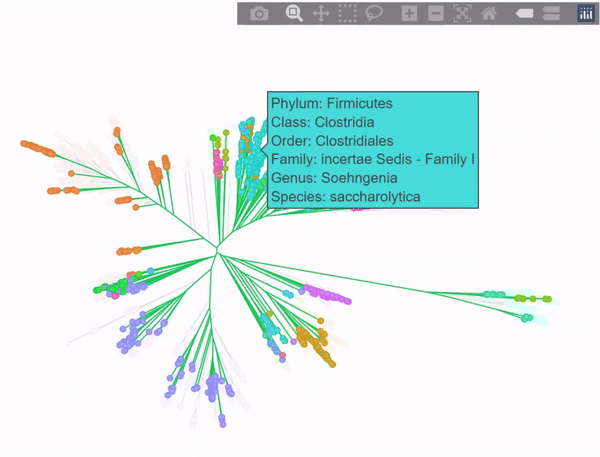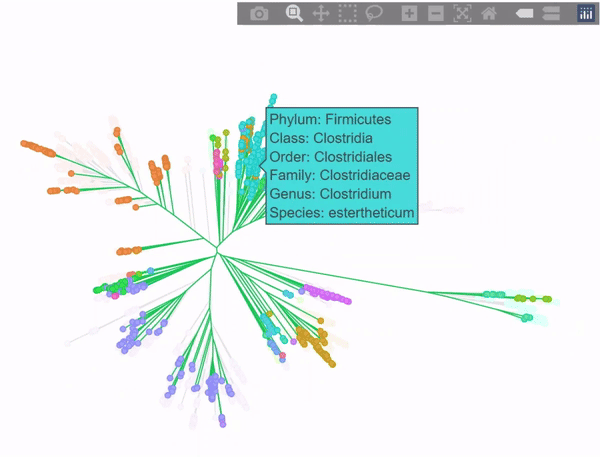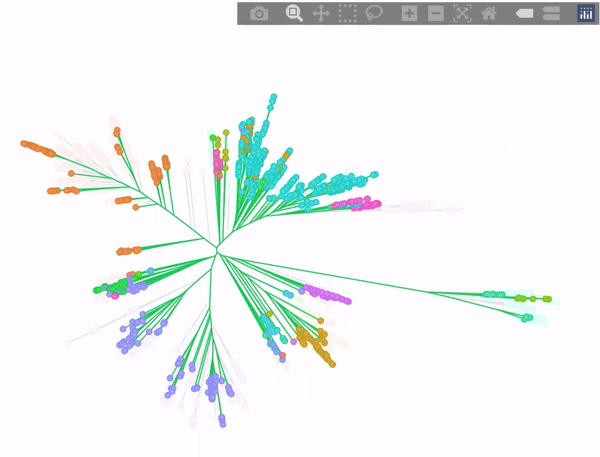
Phylogenetic tree
Shiny app: https://timothy-hackmann.shinyapps.io/FermentationExplorer/
Repo: GitHub - thackmann/FermentationExplorer: 🧪A resource for showing the incredible diversity of fermentative metabolism
Thumbnail:
Full image: