Hi all,
I hope someone could help me with this task.
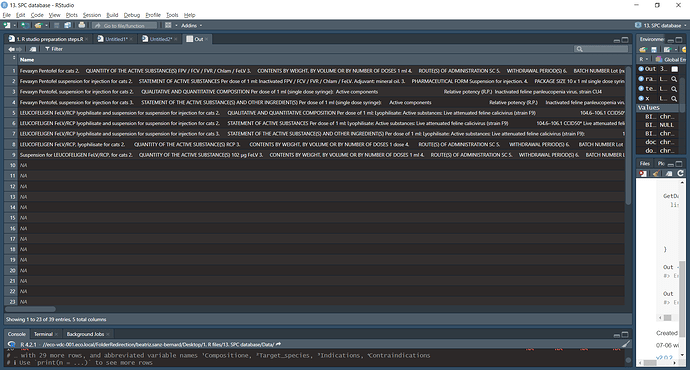
I need to extract from a series of PDF files data contained in sections.
This is the first time I work with PDF text, and after reading on library(pdftools) and regex (of which I never heard before) and checking different examples around, I am going in circles as yet I don't have enough knowledge to get what I need from those examples.
These are some examples of PDF documents that I need to use.
I have tried to download them but they appear with no text, so i opted for saving them in the working directory instead, and attached the file to this message.
Leucofeligen.pdf (154.2 KB)
Pentofel.pdf (207.9 KB)
What I tried to do under GetData is to extract the text following each of the headings of interest. SO far I have detected two issues (although they may be more):
- The headings are repeated in some cases, so multiple entries are generated, while I only want one.
- I don't know how to say what is the end of the text I am interested in. I have used the examples from other regex coding, but they don't work properly.
The ultimate goal is to generate a dataframe with columns as labelled under the GetData function (Name, Composition, target_species, etc.).
I hope the message is clear enough to get some tips...
Many thanks in advance
Beatriz
This is the code I sued in my session with the file I generated after out. Sorry the reprex is underneath but is not working for some reason.
library(pdftools)
#download.file("https://medicines.health.europa.eu/veterinary/en/documents/download/e7da093e-0dc2-4120-8fa1-4beb29cc2a90","Pentofelb.pdf")
#download.file("https://medicines.health.europa.eu/veterinary/en/documents/download/e7da093e-0dc2-4120-8fa1-4beb29cc2a90", "Leucofeligen.pdf")
file.list <- list.files(pattern = "pdf|PDF$")
x <- map(file.list, ~ pdf_text(.))
names(x) <- gsub("\\.pdf", "", file.list)
GetData <- function(x){
list(Name = str_trim(str_extract(x, "(?<=NAME OF THE VETERINARY MEDICINAL PRODUCT)[^-]+")),
Compositione = str_trim(str_extract(x, "(?<=QUALITATIVE AND QUANTITATIVE COMPOSITION)[^-]+")),
Target_species = str_trim(str_extract(x, "(?<=Target species)[^-]+")),
Indications = str_trim(str_extract(x, "(?<=Indications for use, specifying the target species)[^-]+")),
Contraindications = str_trim(str_extract(x, "(?<=Contraindications)[^-]+")))
}
Out <- map_dfr(x, GetData)
Out
The reprex file is this one underneath but is not working - Not sure why is not finding the source documents on my working directory.
#Example
#Download the PDF files.
#download.file("https://medicines.health.europa.eu/veterinary/en/documents/download/e7da093e-0dc2-4120-8fa1-4beb29cc2a90","Pentofelb.pdf")
#download.file("https://medicines.health.europa.eu/veterinary/en/documents/download/e7da093e-0dc2-4120-8fa1-4beb29cc2a90", "Leucofeligen.pdf")
library(pdftools)
#> Warning: package 'pdftools' was built under R version 4.2.3
#> Using poppler version 22.04.0
file.list <- list.files(pattern = "pdf|PDF$")
x <- map(file.list, ~ pdf_text(.))
#> Error in map(file.list, ~pdf_text(.)): could not find function "map"
names(x) <- gsub("\\.pdf", "", file.list)
#> Error in names(x) <- gsub("\\.pdf", "", file.list): object 'x' not found
GetData <- function(x){
list(Name = str_trim(str_extract(x, "(?<=NAME OF THE VETERINARY MEDICINAL PRODUCT)[^-]+")),
Compositione = str_trim(str_extract(x, "(?<=QUALITATIVE AND QUANTITATIVE COMPOSITION)[^-]+")),
Target_species = str_trim(str_extract(x, "(?<=Target species)[^-]+")),
Indications = str_trim(str_extract(x, "(?<=Indications for use, specifying the target species)[^-]+")),
Contraindications = str_trim(str_extract(x, "(?<=Contraindications)[^-]+")))
}
Out <- map_dfr(x, GetData)
#> Error in map_dfr(x, GetData): could not find function "map_dfr"
Out
#> Error in eval(expr, envir, enclos): object 'Out' not found
Created on 2023-07-06 with reprex v2.0.2