Hi,
there is a fairly old post dated November 23, 2016, 18:20, [https://support.rstudio.com/hc/en-us/articles/202133558-Extending-RStudio-with-the-Viewer-Pane], together with a few comments and answers - and I still have the same problem described in it.
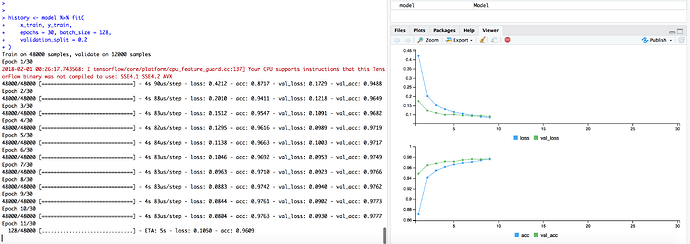
The background: I am working on a neural network in kerasR. There is an existing model and it works as expected within RStudio. The part of the program where my problem arises is the training & validation process. It looks as follows:
history <- model %>% fit(
x_train,
y_train,
batch_size = Bs,
initial_epoch =0,
epochs = trep,
verbose = 2,
view_metrics = "auto",
validation_data = list(x_val, y_val),
shuffle = "batch"
)
As soon as the code suggested in the post mentioned above,
viewer <- getOption("viewer")
viewer("http://localhost:8000")
is inserted right before the line "history..." and the program is run, a message "HTTP Error 404..." appears and nothing else happens in the viewer window. If I click the button "view in new window", a new window opens in my browser, showing in real time what's going on in my application - exactly the same behavior as without the additional code. This is not only rather unpractical, since I would prefer just to look simultaneously on a listing of my ongoing process in parallel to the developing picture within the viewer pane; I also have noticed that the browser produces problems (due to limiting storage space, I suppose) when keeping several (>5) of these windows for later comparison.
To me it looks as if the address given in the second line
viewer("http://localhost:8000")
simply doesn't exist. But I have no idea as to what I could use instead of this one. Any other number instead of the '8000' produces the same result anyway. Do I have to define this address beforehand? And if so - how and where can I do that?
Just in case this might be helpful: I'm using RStudio Version 1.1.383 together with R 3.4.3.
Is this problem really still unsolved? You support would be appreciated - thanks in advance.
Leo