Hello everybody,
Could someone help me in this problem
I’m working on clustering a data and after i got the clustering results , I would like to export each cluster data on a single "csv" file is there a simple and efficient way to do It in this case.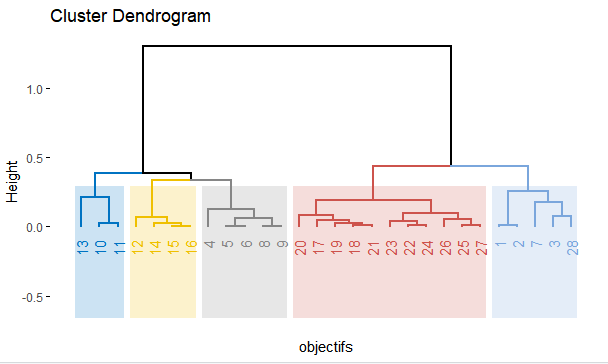
Please post a reproducible example (reprex) of the code you use to make the Dendrogram. Be sure to include the library() calls for the packages you use.
library(leaflet)
library(sp)
mydata<-read.csv("new.csv")
View(mydata)
mydata<-mydata[complete.cases(mydata),]
tableau <- data.frame(x=mydata[,7],y=mydata[,8]);plot(tableau)
mydi = dist(tableau[,],method= "euclidean")
myclust <- hclust(mydi, method="ward.D2")
library(factoextra)
library(ggsci)
require("ggsci")
library(grDevices)
require("grDevices")
fviz_dend(x=myclust,cex = 0.8, lwd = 0.8, k = 5,
k_colors = "jco",
rect = TRUE,
rect_border = "jco",
rect_fill = TRUE,
color_labels_by_k = TRUE,
xlab="objectifs",
main = "Cluster Dendrogram",)
this is the code I've used to generate the dendogram
Can you make mydata reproducible?
The screenshot doesn't help.
It should be reproducible. Like noted in the article posted by @FJCC
People are more likely to answer your question if they can play with the data.
my approach is basically a hack as I'm not practiced with clustering packages and methods to know the 'proper' way to do it. but if you are desperate...this might not be wrong ?
library(tidyverse)
library(factoextra)
library(glue)
mydata <- group_by(iris,Species,.drop=FALSE) %>% group_modify(~head(.)) %>% bind_rows()
mydata<-mydata[complete.cases(mydata),]
tableau <- data.frame(x=mydata[,1],y=mydata[,5]);plot(tableau)
mydi = dist(tableau[,],method= "euclidean")
myclust <- hclust(mydi, method="ward.D2")
(fv <- fviz_dend(x=myclust,cex = 0.8, lwd = 0.8, k = 5,
k_colors = "jco",
rect = TRUE,
rect_border = "jco",
rect_fill = TRUE,
color_labels_by_k = TRUE,
xlab="objectifs",
main = "Cluster Dendrogram",) )
tableau$labels <- fv$plot_env$data$labels$label
tableau$col <- fv$plot_env$data$labels$col
tableau<- mutate(tableau,
clust = dense_rank(col))
walk(unique(tableau$clust),
~write_csv(x = filter(tableau,clust==.),
path = glue("clust{.}.csv")))I don't know if this part of the data like this below is enoughand could help now or not yet
> mydata<-read.csv("new.csv")
> head(mydata)
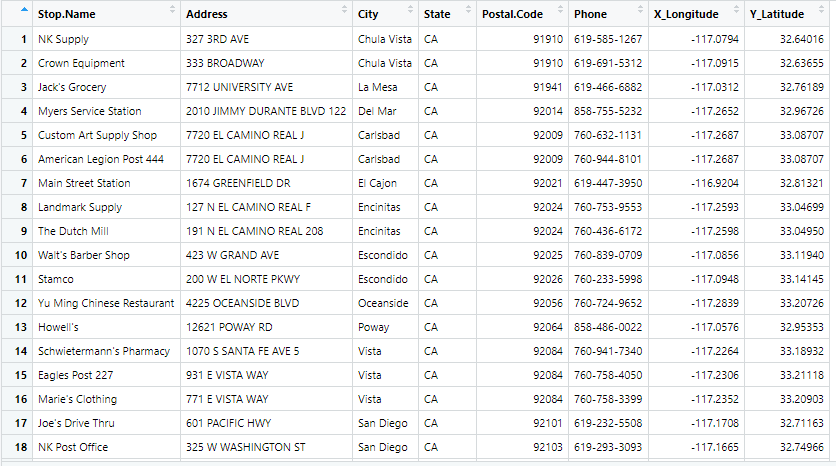
Stop.Name Address City State Postal.Code Phone X_Longitude Y_Latitude
1 NK Supply 327 3RD AVE Chula Vista CA 91910 619-585-1267 -117.0794 32.64016
2 Crown Equipment 333 BROADWAY Chula Vista CA 91910 619-691-5312 -117.0915 32.63655
3 Jack's Grocery 7712 UNIVERSITY AVE La Mesa CA 91941 619-466-6882 -117.0312 32.76189
4 Myers Service Station 2010 JIMMY DURANTE BLVD 122 Del Mar CA 92014 858-755-5232 -117.2652 32.96726
5 Custom Art Supply Shop 7720 EL CAMINO REAL J Carlsbad CA 92009 760-632-1131 -117.2687 33.08707
6 American Legion Post 444 7720 EL CAMINO REAL J Carlsbad CA 92009 760-944-8101 -117.2687 33.08707
This is what it returns
Error in group_by(iris, Species, drop = FALSE) %>% group_modify(~head(.)) %>% :
could not find function "%>%"
> mydata<-mydata[complete.cases(mydata),]
> tableau <- data.frame(x=mydata[,1],y=mydata[,5]);plot(tableau)
Error in plot.window(...) : need finite 'xlim' values
In addition: Warning messages:
1: In xy.coords(x, y, xlabel, ylabel, log) : NAs introduced by coercion
2: In min(x) : no non-missing arguments to min; returning Inf
3: In max(x) : no non-missing arguments to max; returning -Inf
> mydi = dist(tableau[,],method= "euclidean")
Warning message:
In dist(tableau[, ], method = "euclidean") : NAs introduced by coercion
> myclust <- hclust(mydi, method="ward.D2")
>
>
> (fv <- fviz_dend(x=myclust,cex = 0.8, lwd = 0.8, k = 5,
+ k_colors = "jco",
+ rect = TRUE,
+ rect_border = "jco",
+ rect_fill = TRUE,
+ color_labels_by_k = TRUE,
+ xlab="objectifs",
+ main = "Cluster Dendrogram",) )
>
> tableau$labels <- fv$plot_env$data$labels$label
> tableau$col <- fv$plot_env$data$labels$col
>
>
> tableau<- mutate(tableau,
+ clust = dense_rank(col))
Error in mutate(tableau, clust = dense_rank(col)) :
could not find function "mutate"
>
> walk(unique(tableau$clust),
+ ~write_csv(x = filter(tableau,clust==.),
+ path = glue("clust{.}.csv")))
Error in walk(unique(tableau$clust), ~write_csv(x = filter(tableau, clust == :
could not find function "walk"
the magrittr pipe is part of the tidyverse.
can you load the tidyverse library ?
what is the result of
(require(tidyverse))
?
this is the new results generated
> library(tidyverse)
> library(factoextra)
> library(glue)
> #install.packages("tidyverse")
> library(tidyverse)
> require("tidyverse")
> mydata<-read.csv("new.csv")
> mydata <- group_by(iris,Species,drop=FALSE) %>% group_modify(~head(.)) %>% bind_rows()
> mydata<-mydata[complete.cases(mydata),]
> tableau <- data.frame(x=mydata[,1],y=mydata[,5]);plot(tableau)
> mydi = dist(tableau[,],method= "euclidean")
Warning message:
In dist(tableau[, ], method = "euclidean") : NAs introduced by coercion
> myclust <- hclust(mydi, method="ward.D2")
>
>
> (fv <- fviz_dend(x=myclust,cex = 0.8, lwd = 0.8, k = 5,
+ k_colors = "jco",
+ rect = TRUE,
+ rect_border = "jco",
+ rect_fill = TRUE,
+ color_labels_by_k = TRUE,
+ xlab="objectifs",
+ main = "Cluster Dendrogram",) )
>
> tableau$labels <- fv$plot_env$data$labels$label
> tableau$col <- fv$plot_env$data$labels$col
>
>
> tableau<- mutate(tableau,
+ clust = dense_rank(col))
>
> walk(unique(tableau$clust),
+ ~write_csv(x = filter(tableau,clust==.),
+ path = glue("clust{.}.csv")))
Yes, and ?...
All good?
I'm not sure because I don't know exactly where the clusters files should be generated after this compilation
In your working directory...
You can have R tell you where that is
getwd()I think the the foldels generated contain the individuels ID but not contain the rest of informations attributed to them on the original data you can see the screens below .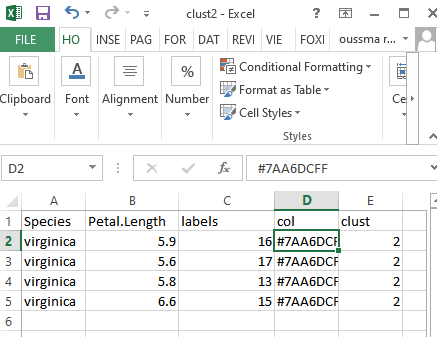 !
!
Its made from tablaeu...
this is a part of the data
> mydata<-read.csv("new.csv")
> head(mydata)
Stop.Name Address City State Postal.Code Phone X_Longitude Y_Latitude
1 NK Supply 327 3RD AVE Chula Vista CA 91910 619-585-1267 -117.0794 32.64016
2 Crown Equipment 333 BROADWAY Chula Vista CA 91910 619-691-5312 -117.0915 32.63655
3 Jack's Grocery 7712 UNIVERSITY AVE La Mesa CA 91941 619-466-6882 -117.0312 32.76189
4 Myers Service Station 2010 JIMMY DURANTE BLVD 122 Del Mar CA 92014 858-755-5232 -117.2652 32.96726
5 Custom Art Supply Shop 7720 EL CAMINO REAL J Carlsbad CA 92009 760-632-1131 -117.2687 33.08707
6 American Legion Post 444 7720 EL CAMINO REAL J Carlsbad CA 92009 760-944-8101 -117.2687 33.08707
so perhaps change the parts of my solution involving tableau to involve mydata, if thats what you prefer
yeah anyway thank you so much I'll work on what you said
This topic was automatically closed 21 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.