Please ask your questions about Quarto or R Markdown here.
I keep getting this message when I knit my Markdown file:
Quitting from Bellabeat-Capstone-Project.Rmd:57-67 [Importing data frames]
Execution halted
I have pasted the lines 57-67 below.
At first the Error message I got was: error: ! object 'dailyActivity_merged' not found Backtrace: ▆ 1. └─janitor::clean_names(dailyActivity_merged) Quitting from Bellabeat-Capstone-Project.Rmd:61-70 [Clean Names] Execution halted
Now execution is just halted
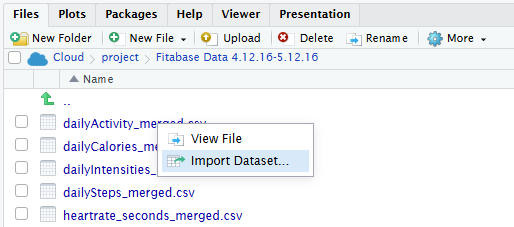
the 'path_to_file' insertion is from a previous user who faced a similar problem and he inserted 'path_to_file' which worked. I just don't know where he inserted it.
And I can't find the post
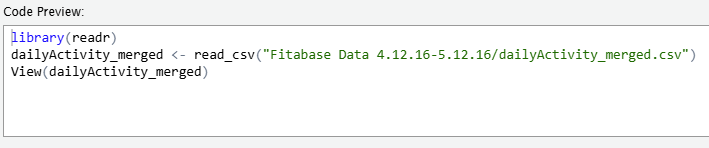
dailyActivity_merged <-path_to_file, read_csv('/cloud/project/mturkfitbit_export_4.12.16-5.12.16/Fitabase Data 4.12.16-5.12.16_dailyActivity_merged.csv)
dailyIntensities_merged <- path_to_file, read_csv('/cloud/project/mturkfitbit_export_4.12.16-5.12.16/Fitabase Data 4.12.16-5.12.16_dailyIntensities_merged.csv)
dailySteps_merged <- path_to_file, read_csv('/cloud/project/mturkfitbit_export_4.12.16-5.12.16/Fitabase Data 4.12.16-5.12.16_dailySteps_merged.csv)
dailyCalories_merged <- path_to_file, read_csv('/cloud/project/mturkfitbit_export_4.12.16-5.12.16/Fitabase Data 4.12.16-5.12.16_dailyCalories_merged.csv)
sleepDay_merged <- path_to_file, read_csv('/cloud/project/mturkfitbit_export_4.12.16-5.12.16/Fitabase Data 4.12.16-5.12.16_sleepDay_merged.csv)