I'm experiencing an error message when working with a list column after using rsample. I created a sample of sanitized data as a csv and it seems to reproduce the issue. This csv file here.
(Incidentally is there a better way to share example data on here?)
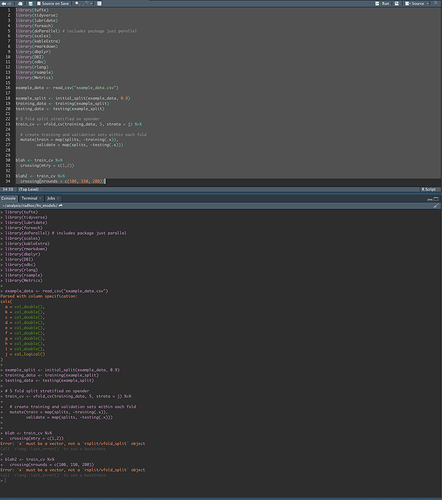
Here is the script and error message:
# (so you know what's loaded in my environment)
library(tufte)
library(tidyverse)
library(lubridate)
library(foreach)
library(doParallel)
library(scales)
library(kableExtra)
library(rmarkdown)
library(dbplyr)
library(DBI)
library(odbc)
library(rlang)
library(rsample)
library(Metrics)
example_data <- read_csv("example_data.csv")
example_split <- initial_split(example_data, 0.9)
training_data <- training(example_split)
testing_data <- testing(example_split)
# 5 fold split stratified on spender
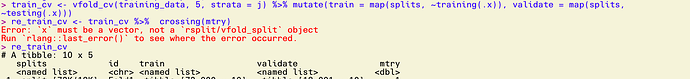
train_cv <- vfold_cv(training_data, 5, strata = j) %>%
# create training and validation sets within each fold
mutate(train = map(splits, ~training(.x)),
validate = map(splits, ~testing(.x)))
# everything works up till this point. It's when I try to do anything with train_cv that I encounter issues.
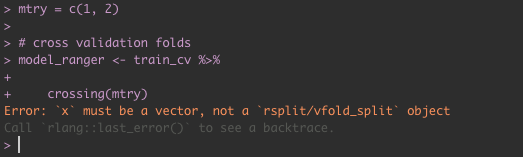
blah <- train_cv %>%
crossing(mtry = c(1,2))
> Error: `x` must be a vector, not a `rsplit/vfold_split` object
blah2 <- train_cv %>%
crossing(nrounds = c(100, 150, 200))
> Error: `x` must be a vector, not a `rsplit/vfold_split` object
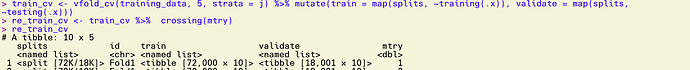
Here's how train_cv looks before trying to use it with crossing():
train_cv
# 5-fold cross-validation using stratification
# A tibble: 5 x 4
splits id train validate
* <named list> <chr> <named list> <named list>
1 <split [72K/18K]> Fold1 <tibble [72,000 × 10]> <tibble [18,001 × 10]>
2 <split [72K/18K]> Fold2 <tibble [72,001 × 10]> <tibble [18,000 × 10]>
3 <split [72K/18K]> Fold3 <tibble [72,001 × 10]> <tibble [18,000 × 10]>
4 <split [72K/18K]> Fold4 <tibble [72,001 × 10]> <tibble [18,000 × 10]>
5 <split [72K/18K]> Fold5 <tibble [72,001 × 10]> <tibble [18,000 × 10]>
Desired outcome is that there will be a new column 'mtry' and each existing row (fold) will have a row for mtry = 1 and another for mtry = 2.
How can I continue to work with train_cv after creating folds and then use crossing to experiment with various hyper parameters in my workflow?
[EDIT]
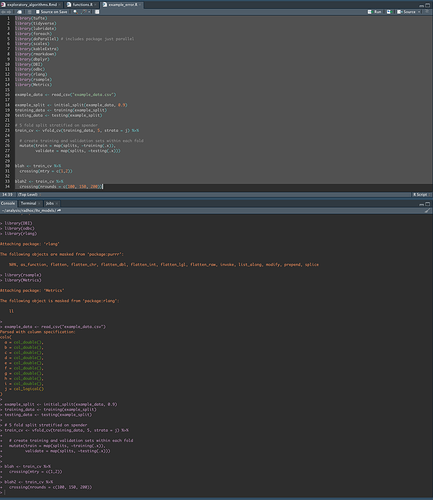
This error is inconsistent. I tried restarting my session and it ran fine. But in my script I cannot restart my session each time I come to this code block. What could be loaded in my space that would lead to this error? Screen shot:
And then after clearing workspace and starting a fresh session, everything works:
In case it's informative, here's what I'm shown when I click 'show traceback' on the error message:
Error: `x` must be a vector, not a `rsplit/vfold_split` object
23.
stop(fallback)
22.
signal_abort(cnd)
21.
abort(message, .subclass = c(.subclass, "vctrs_error"), ...)
20.
stop_vctrs(msg, "vctrs_error_scalar_type", actual = x)
19.
stop_scalar_type(.Primitive("quote")(structure(list(data = structure(list( s = c("92DF3481-4F83-47E4-AE08-E7AD35EBC2B9", "IDFV-DB587A66-50ED-4468-999D-81CB9D872B81", "EAB6422C-17D7-428C-B25A-9BA9EB5C6FE2", "IDFV-A228265A-CB20-40EE-BEFF-85A532525DA2", "IDFV-109FD148-8287-47BF-A301-130557CC2583", "6B611B4F-6C1B-45D2-99E7-3BA9BF67CC27", ...
18.
vec_unique_loc(x)
17.
vec_slice(x, vec_unique_loc(x))
16.
vec_unique(x)
15.
vec_proxy_compare(x)
14.
is.data.frame(proxy)
13.
order_proxy(vec_proxy_compare(x), direction = direction, na_value = na_value)
12.
vec_order(x, direction = direction, na_value = na_value)
11.
vec_sort(vec_unique(x))
10.
.f(.x[[i]], ...)
9.
map(cols, sorted_unique)
8.
crossing(., mtry = c(1, 2))
7.
function_list[[i]](value)
6.
freduce(value, `_function_list`)
5.
`_fseq`(`_lhs`)
4.
eval(quote(`_fseq`(`_lhs`)), env, env)
3.
eval(quote(`_fseq`(`_lhs`)), env, env)
2.
withVisible(eval(quote(`_fseq`(`_lhs`)), env, env))
1.
train_cv %>% crossing(mtry = c(1, 2)) %>% mutate(model_binary = map2(.x = train, .y = mtry, ~ranger::ranger(formula = spender ~ d7_utility_sum + recent_utility_ratio, probability = T, mtry = .y, data = .x %>% filter(spend_7d == 0) %>% mutate(spender = factor(spender)))), ...