Hi!
Last week I read in some data in R to learn to work with R studio for my thesis.
It all worked back then, however I wanted to work further with the same data so i saved the code.
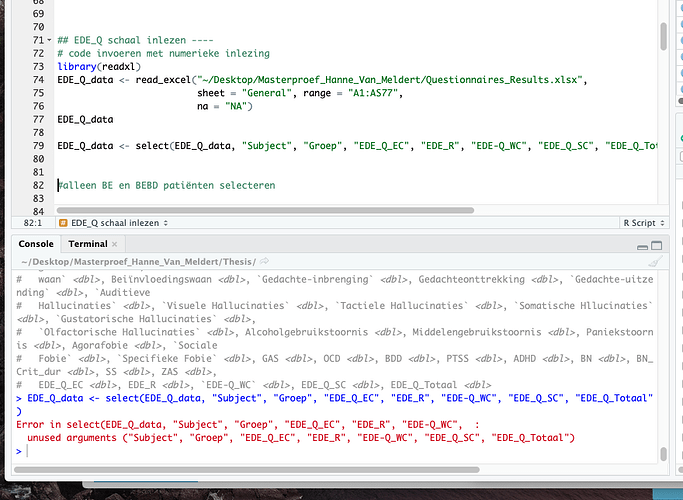
No I wanted to work further, but suddenly get ERRORS when using the 'select' function (although last week i did not give any ERRORS for the same data)
I will upload a photo with the error.
Does someone see what I doe wrong?
Thanks!
if you want to use select function from the dplyr package of the tidyverse then you should use library(tidyverse) before calling select.
Ooh sorry, guess I overlooked that I didn't do that!
Thanks!
So, I tried it again after you reply and it worked, now today I suddenly get the same error, even while I used the library(tidyverse)...I don't get what I'm doing wrong?

please provide a reprex
FAQ: How to do a minimal reproducible example ( reprex ) for beginners
please slow down and take the time to read the linked document on how to reprex all through before jumping ahead. Images are not a good way to share information. I appreciate your enthusiasm, but learning forum ettiquet and good practices for debugging and sharing information with peers will pay you big dividens down the road on your journey to being a confident R user. ![]()
Will try to post it in the wright form!
data.frame(
stringsAsFactors = FALSE,
check.names = FALSE,
Sub = c(21, 31, 33, 44, 47, 48),
Groep = c("BE","BE",
"BEBD","BE","BE","BE"),
Behandeling = c(1, 1, 1, 1, 1, 1),
`1.1` = c(3, 4, 4, 3, 5, 5),
`1.2` = c(5, 5, 5, 5, 4, 2),
`1.3` = c(5, 5, 5, 5, 5, 4),
`1.4` = c(3, 2, 4, 3, 1, 1),
`1.5` = c(5, 5, 5, 5, 4, 4),
`1.6` = c(5, 5, 2, 5, 3, 5))
#> Sub Groep Behandeling 1.1 1.2 1.3 1.4 1.5 1.6
#> 1 21 BE 1 3 5 5 3 5 5
#> 2 31 BE 1 4 5 5 2 5 5
#> 3 33 BEBD 1 4 5 5 4 5 2
#> 4 44 BE 1 3 5 5 3 5 5
#> 5 47 BE 1 5 4 5 1 4 3
#> 6 48 BE 1 5 2 4 1 4 5
Created on 2020-06-17 by the reprex package (v0.3.0)
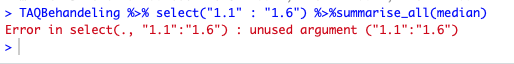
TAQBehandeling %>% select("1.1" : "1.6") %>%summarise_all(median)
Thank you,
My first comment is that in a fresh session the code works.
library(tidyverse)
df<- data.frame(
stringsAsFactors = FALSE,
check.names = FALSE,
Sub = c(21, 31, 33, 44, 47, 48),
Groep = c("BE","BE",
"BEBD","BE","BE","BE"),
Behandeling = c(1, 1, 1, 1, 1, 1),
`1.1` = c(3, 4, 4, 3, 5, 5),
`1.2` = c(5, 5, 5, 5, 4, 2),
`1.3` = c(5, 5, 5, 5, 5, 4),
`1.4` = c(3, 2, 4, 3, 1, 1),
`1.5` = c(5, 5, 5, 5, 4, 4),
`1.6` = c(5, 5, 2, 5, 3, 5))
df %>% select("1.1" : "1.6") %>%summarise_all(median)
do you load other packages with conflicting definitions of select ?
what result do you get in the console when you type
getAnywhere(select)getAnywhere(select)
no object named ‘select’ was found
I used these packages library(ordinal)
library(dplyr)
library(xlsx)
library(Hmisc)
library(xlsx2dfs)
library(readxl)
library(openxlsx)
library(tidyverse)
library(datapasta)
library(reprex)
thats very strange, I dont know whats wrong with your system.
When I do it I get
> getAnywhere(select)
2 differing objects matching ‘select’ were found
in the following places
package:plotly
package:dplyr
namespace:tidyselect
namespace:dplyr
what about if you change your code to say dplyr::select instead of select on its own ?
getAnywhere(dplyr::select)
A single object matching ‘::’ ‘dplyr’ ‘select’ was found
It was found in the following places
package:base
namespace:base
with value
function (pkg, name)
{
pkg <- as.character(substitute(pkg))
name <- as.character(substitute(name))
getExportedValue(pkg, name)
}
<bytecode: 0x106831998>
<environment: namespace:base>
Warning message:
In find(x, numeric = TRUE) :
elements of 'what' after the first will be ignored
sorry no.
getAnywhere(select) would be correct. and if library(dplyr) really had been loaded it should be mentioned there.
I was suggesting being explicit about dplyr in the actual code
df %>% dplyr::select("1.1" : "1.6") %>%summarise_all(median)
though this should be uneccasry if you really have dplyr and really loaded it with either library(dplyr) or require(dplyr)
When you require(dplyr) what shows on the console ?
and if you getAnywhere(select) immediately after that ?
Hi Nirgrahamuk!
After not loading all the packages in R I needed further in my calculations, the error did not occur.
So I guess some of those packages conflicted like you said...
Thanks!
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.