Amalie
August 31, 2022, 8:50am
1
Hi can someone please help me with the following error msg - I think it's due to the colonne Fat_protein isn't nummeric and that's why I have tried to add the as.factor(Mouse$Fat_Protein) but i still get the same error.
Mouse <- read_excel('Mouse_diet_intervention.xlsx')
Mouse <- Mouse[c(1:40),]
Mouse$Fat_Protein <- as.factor(Mouse$Fat_Protein)
Mouse_m <- Mouse [,c(5,23:28)]
MousePCA <- prcomp(Mouse_m, center=TRUE, scale=TRUE)
Error msg:
Error in colMeans(x, na.rm = TRUE) : 'x' must be numeric
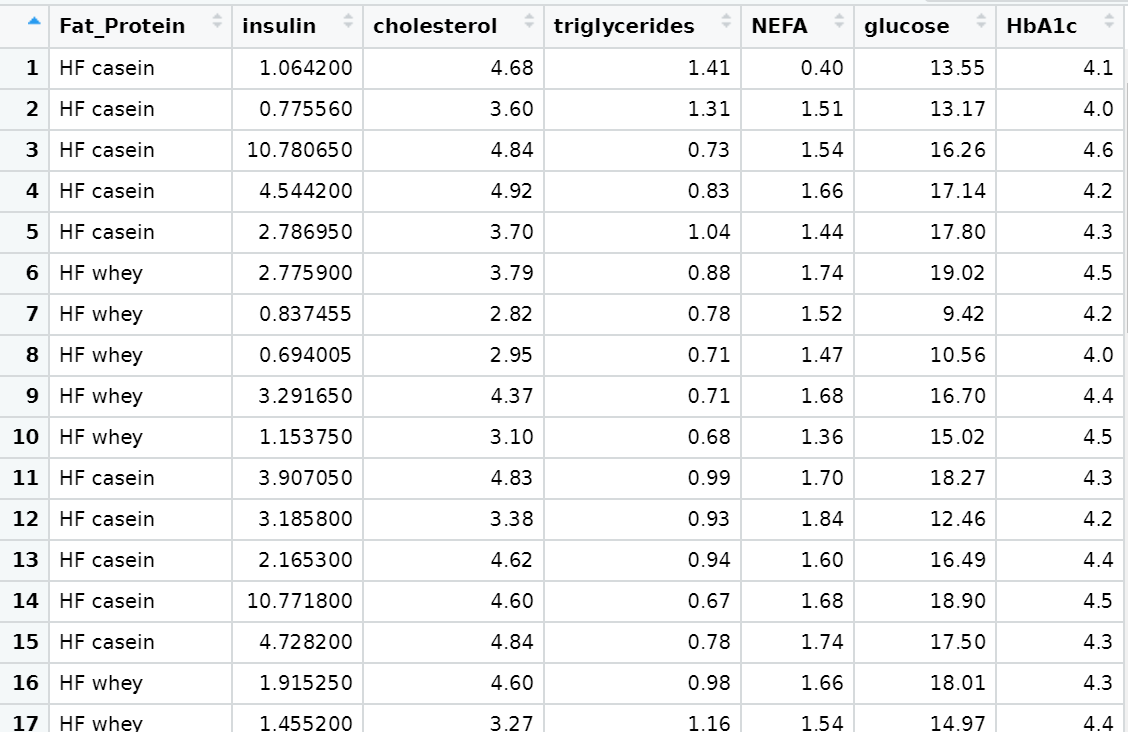
My data for Mouse_m:
Amalie
August 31, 2022, 10:29am
4
So I'm about to have a course in R after the summerbreak where we have recieved some tasks we could do before the course starts in order not to be totally lost. So this one it's says "Suggest two ways in which you can change the coding so that the qplot can be made" where I got the code from the task to begin with - so I just added the Mouse$Fat_Protein <- as.factor() because I thought the error showed due to that colonne since it wasn't numeric but it didn't solve the issue
Sorry about my english btw it's not my first language
factors are not directly numeric; they are integers underneath yes, but you wont have access to that aspect of them without using as.integer()
Amalie
August 31, 2022, 1:25pm
6
Thanks! I have tried to use as.integer(Mouse$Fat_Protein) but it still don't work
For make prcomp() you need all data like numeric or in specific case put the factor variables. But don't put character variables.
Send you a great guide for this:
we cant see your code to judge how you did it. but maybe this example will help you.
df_ <- data.frame(
a=letters[1:2],
b=1:2
)
prcomp(df_ )
df_$a <- as.integer(as.factor(df_$a))
prcomp(df_ )
Amalie
August 31, 2022, 2:05pm
9
Thanks I have typed in the following code:
Mouse <- read_excel('Mouse_diet_intervention.xlsx')
Mouse <- Mouse[c(1:40),]
Mouse_m <- Mouse [,c(5,23:28)]
Mouse$Fat_Protein <- as.integer(as.factor(Mouse$Fat_Protein))
MousePCA <- prcomp(Mouse_m, center=TRUE, scale=TRUE)
But I still get the same error:
Error in colMeans(x, na.rm = TRUE) : 'x' must be numeric
Did I type it in wrong?
For better help you all the community, put a reproducible example of data.
A minimal reproducible example consists of the following items:
A minimal dataset, necessary to reproduce the issue
The minimal runnable code necessary to reproduce the issue, which can be run
on the given dataset, and including the necessary information on the used packages.
Let's quickly go over each one of these with examples:
Like this:
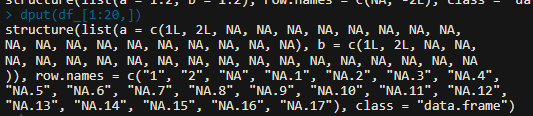
dput(Mouse[1:30 , ]
copy and paste the result of this code.
Amalie
August 31, 2022, 2:27pm
11
Thanks where in the code should i inset in?
Yes, the result in the console. Copy an paste the result of dput(Mouse[1:30 , ].
Amalie:
Mouse_m <- Mouse [,c(5,23:28)]
Mouse$Fat_Protein <- as.integer(as.factor(Mouse$Fat_Protein))
MousePCA <- prcomp(Mouse_m, center=TRUE, scale=TRUE)
This shows you adjust Mouse, not Mouse_m to have the integer represntation of fat protein,
system
September 7, 2022, 2:44pm
15
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.