Hello,
Back with yet another challenge that I can't seem to figure out. Again, I'm very new to all of this, so I'm not sure if I'm even explaining my problem clearly below, but I'll try.
I want to plot points on a world map (particularly Brazil) using ggplot2. The points represent zip codes across Brazil. I followed a YouTube video to get the code I think I need; however, I'm getting the following error:
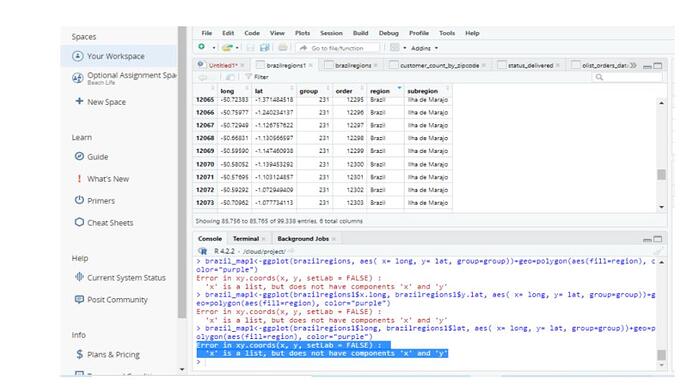
Error in xy.coords(x, y, setLab = FALSE) :
'x' is a list, but does not have components 'x' and 'y'
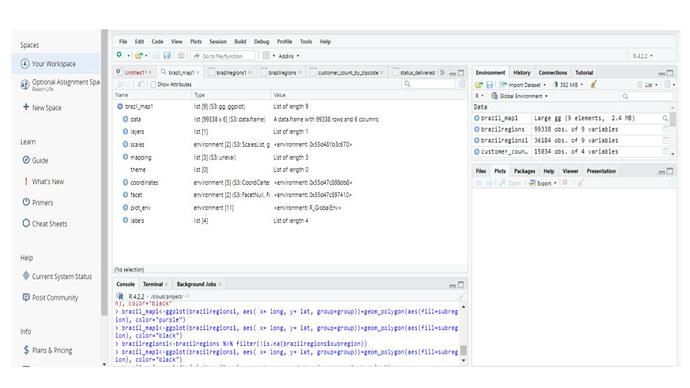
Here is the df I'm working with:
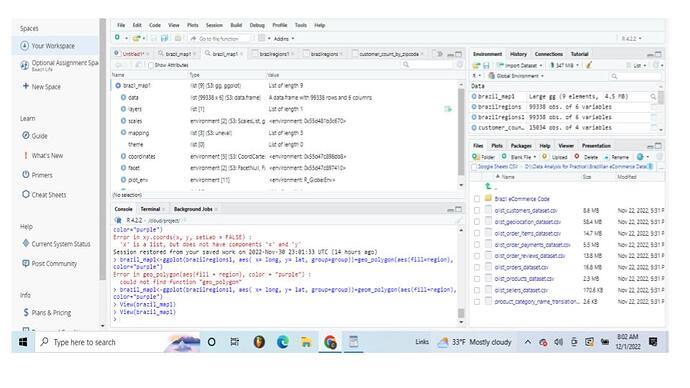
I created a df called brazilregions using map_data
(brazilregions<-map_data("world")
I then joined my original df with this new df:
(brazilregions<-left_join(brazilregions,customer_count_by_zipcode,by="regions)
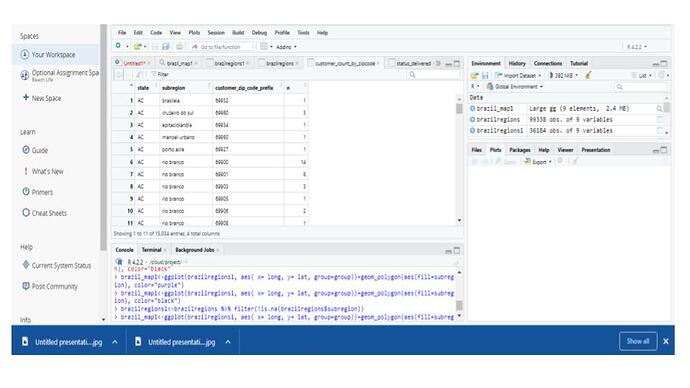
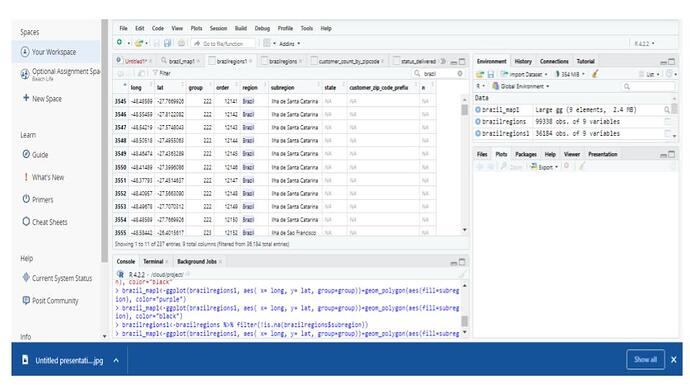
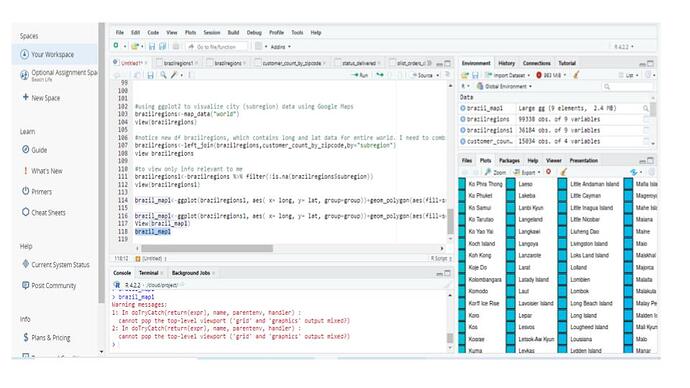
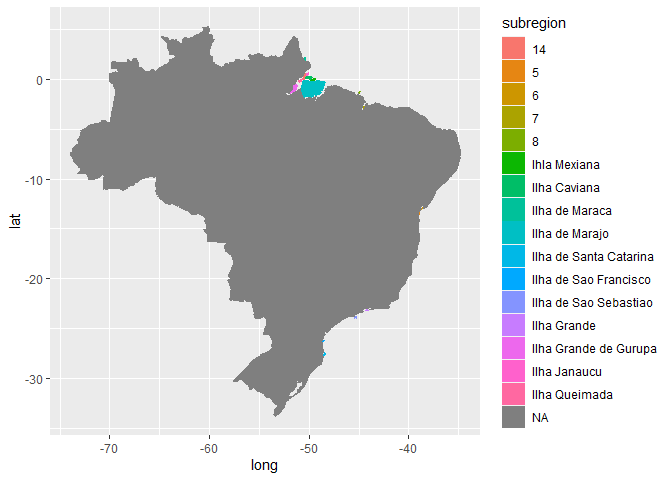
I added a filter to view only the info relevant to me. However, my list still contains ALL of the map_data info as before. It's just that now, SOME of my Brazil info (states in Brazil) are populated, but not all. There are still a ton of NAs.
brazilregions1<-brazilregions %>% filter(!is.na(brazilregions$region))
Using the code as explained in the YouTube video, I constructed this:
brazil_map1<-ggplot(brazilregions1, aes( x= long, y= lat, group=group))+geo=polygon(aes(fill=region), color="purple")
But got this message:
Error in xy.coords(x, y, setLab = FALSE) : 'x' is a list, but does not have components 'x' and 'y'
I don't know if this is enough information for anyone to go on to offer any feedback on what I should do but this is what I have. If there is something in particular I'm leaving out that would help you help me, please let me know. Any suggestion is greatly appreciated.
PS: Here is the link to the YT video I used as reference: https://www.youtube.com/watch?v=AgWgPSZ7Gp0