Is this what you mean?
library(ggplot2)
library(viridis)
library(scales)
# Sample data on a copy/paste friendly format, replace with your actual data frame.
matchedsaltfixed <- data.frame(
stringsAsFactors = FALSE,
F1 = c("f1mock","f1mock","f1mock",
"f1mock","f1mock","f1mock","f1mock","f1mock","f1mock",
"f1mock","f1mock","f1mock","f1mock","f1mock",
"f1mock","f1mock","f1PSTLOW","f1PSTLOW","f1PSTLOW",
"f1PSTLOW","f1PSTLOW","f1PSTLOW","f1PSTLOW","f1PSTLOW",
"f1PSTLOW","f1PSTLOW","f1PSTLOW","f1PSTLOW","f1PSTLOW",
"f1PSTLOW","f1PSTLOW","f1PSTLOW","f1PSTMED",
"f1PSTMED","f1PSTMED","f1PSTMED","f1PSTMED","f1PSTMED",
"f1PSTMED","f1PSTMED","f1PSTMED","f1PSTMED","f1PSTMED",
"f1PSTMED","f1PSTMED","f1PSTMED","f1PSTMED","f1PSTMED",
"f1PSTHIGH","f1PSTHIGH","f1PSTHIGH","f1PSTHIGH",
"f1PSTHIGH","f1PSTHIGH","f1PSTHIGH","f1PSTHIGH",
"f1PSTHIGH","f1PSTHIGH","f1PSTHIGH","f1PSTHIGH","f1PSTHIGH",
"f1PSTHIGH","f1PSTHIGH","f1PSTHIGH"),
LineHPA = c("m-1","m-1","m-1","m-1",
"m-2","m-2","m-2","m-2","m-3","m-3","m-3","m-3",
"m-4","m-4","m-4","m-4","Pst-I1","Pst-I1","Pst-I1",
"Pst-I1","Pst-I2","Pst-I2","Pst-I2","Pst-I2","Pst-I3",
"Pst-I3","Pst-I3","Pst-I3","Pst-I4","Pst-I4",
"Pst-I4","Pst-I4","Pst-II1","Pst-II1","Pst-II1","Pst-II1",
"Pst-II2","Pst-II2","Pst-II2","Pst-II2","Pst-I3",
"Pst-I3","Pst-I3","Pst-I3","Pst-II4","Pst-II4",
"Pst-II4","Pst-II4","Pst-III1","Pst-III1","Pst-III1",
"Pst-III1","Pst-III2","Pst-III2","Pst-III2","Pst-III2",
"Pst-III3","Pst-III3","Pst-III3","Pst-III3","Pst-III4",
"Pst-III4","Pst-III4","Pst-III4"),
P = c(1L,2L,3L,4L,1L,2L,3L,4L,
1L,2L,3L,4L,1L,2L,3L,4L,1L,2L,3L,4L,1L,2L,
3L,4L,1L,2L,3L,4L,1L,2L,3L,4L,1L,2L,3L,4L,
1L,2L,3L,4L,1L,2L,3L,4L,1L,2L,3L,4L,1L,2L,3L,
4L,1L,2L,3L,4L,1L,2L,3L,4L,1L,2L,3L,4L),
DAMAGE = c(2L,21L,151L,49L,0L,16L,
151L,62L,6L,30L,163L,42L,8L,14L,147L,40L,10L,49L,
184L,40L,12L,50L,150L,29L,4L,33L,139L,32L,13L,
36L,139L,41L,10L,29L,156L,37L,0L,11L,40L,14L,
2L,21L,61L,17L,2L,41L,132L,43L,20L,40L,147L,
22L,21L,62L,124L,38L,5L,50L,189L,40L,16L,52L,
158L,30L)
)
ggplot(matchedsaltfixed, aes(x = LineHPA, y = DAMAGE, fill = P)) +
geom_col(position = "fill") +
scale_colour_viridis() +
scale_y_continuous(labels = percent) +
labs(x = "Lines - Hpa",
y = "% Leaves") +
facet_wrap(facets = vars(F1), scales = "free_x")
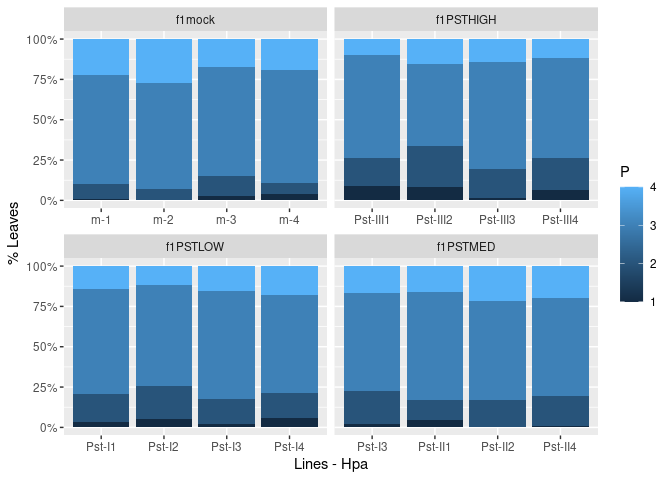
Created on 2021-02-20 by the reprex package (v1.0.0)
BTW, please notice the way I'm sharing the example with you, that would be a proper reproducible example as explained on the guide I linked for you before. Next time please make your questions providing a reprex like this one.