I keep getting the same error - despite checking that the file name is correct and has been imported. Here's my code:
# Setting up the RStudio environment
#~~~~~~~~~~~
# Libraries
#~~~~~~~~~~
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(ggplot2)
#> Registered S3 methods overwritten by 'ggplot2':
#> method from
#> [.quosures rlang
#> c.quosures rlang
#> print.quosures rlang
library(tidyr)
#~~~~~~~~~~~
# Functions
#~~~~~~~~~~
rescale_01 <-function(x) (x-min(x))/(max(x)-min(x)) -1/2
z_stand<-function(x) (x-mean(x))/sd(x)
ExpectedBrix <- function(x) (x*0.21084778699754 + 4.28455310831511)
#~~~~~~~~~~~~
# Thresholds
#~~~~~~~~~~~
Thresh.Brix.min <- 15
Thresh.Brix.max <- 30
Thresh.Pol.min <- 50
Thresh.Pol.max <- 105
ExpectedBrix.delta <- 1
Thresh.Fibre.min <- 4
Thresh.Fibre.max <- 25
Thresh.Fibre.delta <- .25
Thresh.Ash.min <- 0
Thresh.Ash.max <- 8
# Import the Fibre data into a data table called “Lab_Fibre_Data”
Lab_Fibre_Data <- read.table("Lab_Fibre_Weights.csv", header=TRUE, sep=",", dec =".")
#> Warning in file(file, "rt"): cannot open file 'Lab_Fibre_Weights.csv': No
#> such file or directory
#> Error in file(file, "rt"): cannot open the connection
# Calculate Percentage Fibre Variables using direct assignment
# Name the resulting variable “Fibre1” and add this new variable to the “Lab_Fibre_Data” data table, using direct assignment
# Fibre% = 100 * (InitialSampleCanWeight – FinalSampleCanWeight) / SampleWeight
Lab_Fibre_Data$Fibre1 <- 100 * (Lab_Fibre_Data$InitialSampleCanWeight_1 - Lab_Fibre_Data$FinalSampleCanWeight_1) / Lab_Fibre_Data$SampleWeight_1
#> Error in eval(expr, envir, enclos): object 'Lab_Fibre_Data' not found
# Calculate Percentage Fibre Variables using the mutate() function from the dplyr package, call it “Fibre2”
# Add “Fibre2” as a new variable to the “Lab_Fibre_Data” data table
Lab_Fibre_Data <- Lab_Fibre_Data %>% mutate(Fibre2 = 100 *(InitialSampleCanWeight_2 - FinalSampleCanWeight_2) / SampleWeight_2)
#> Error in eval(lhs, parent, parent): object 'Lab_Fibre_Data' not found
# Use the filter() function to remove samples (rows) that contain a missing value
Lab_Fibre_Filtered <- Lab_Fibre_Data %>% filter(InitialSampleCanWeight_1 >0 &
InitialSampleCanWeight_2 >0 &
FinalSampleCanWeight_1 >0 &
FinalSampleCanWeight_2 >0 &
SampleWeight_1 >0 &
SampleWeight_2 >0)
#> Error in eval(lhs, parent, parent): object 'Lab_Fibre_Data' not found
# UPDATE code in table 5 to include maximum fibre difference limit as an additional filtering criteria
# Absolute difference between the two estimates (computed variables “Fibre1” and “Fibre2”) = or > 0.25 units
# UPDATE your code in table 5 to include this maximum fibre difference limit as an additional filtering criteria
Lab_Fibre_Filtered <- Lab_Fibre_Filtered %>% filter(!(abs(Fibre1 - Fibre2) >= 0.25))
#> Error in eval(lhs, parent, parent): object 'Lab_Fibre_Filtered' not found
# Calculate the final fibre estimates by averaging the replicate fibre measurements, “Fibre1” and “Fibre2”
# Use mutate() to calculate the average fibre and add it as a new variable named “Fibre” to the Lab_Fibre_Filtered data table
Lab_Fibre_Filtered <- Lab_Fibre_Filtered %>% mutate(Fibre = (Fibre1 + Fibre2)/2)
#> Error in eval(lhs, parent, parent): object 'Lab_Fibre_Filtered' not found
# Use a PIPE to sequentially filter the measurements in Lab_Fibre_Filtered to remove the out-of-range fibre values
# Keeping only the rows of the data table for which “Fibre” is greater than Thresh.Fibre.min
# Keeping only the resulting rows for which “Fibre” is less than Thresh.Fibre.max. Save the resulting data into the Lab_Fibre_Filtered table
Lab_Fibre_Filtered <- Lab_Fibre_Filtered %>% filter(Fibre > Thresh.Fibre.min & Fibre < Thresh.Fibre.max)
#> Error in eval(lhs, parent, parent): object 'Lab_Fibre_Filtered' not found
# Use select() to save the LabID and Fibre variables (1st and last columns) from Lab_Fibre_Filtered to a new data table called Lab_Fibre
Lab_Fibre <- Lab_Fibre_Filtered %>% select(LabID,Fibre)
#> Error in eval(lhs, parent, parent): object 'Lab_Fibre_Filtered' not found
# Import the Ash data into a data table called “Lab_Ash_Data”
Lab_Ash_Data <- read.table("Lab_Ash_Weights.csv", header=TRUE, sep=",", dec =".")
#> Warning in file(file, "rt"): cannot open file 'Lab_Ash_Weights.csv': No
#> such file or directory
#> Error in file(file, "rt"): cannot open the connection
# Calculate Ash Variables
# Use a PIPE to filter out the missing values using filter()
# Sequentially calculate “InitialWeight”, “FinalWeight” and “Ash” as new variables using mutate()
# TinWeight (column 2), InitialSampleInTinWeight (column 3), FinalSampleInTinWeight (column 4)
# Ash = 100*FinalWeight/InitialWeight
# InitialWeight=InitialSampleInTinWeight-TinWeight, FinalWeight=FinalSampleInTinWeight-TinWeight
Lab_Ash_Calculated$Ash <- Lab_Ash_Data %>%
filter(InitialSampleInTinWeight ==0 &
FinalSampleInTinWeight ==0) &
mutate(InitialWeight = InitialSampleInTinWeight) &
mutate(FinalWeight = FinalSampleInTinWeight) &
mutate(Ash = 100 *(FinalWeight/InitialWeight))
#> Error in eval(lhs, parent, parent): object 'Lab_Ash_Data' not found
# Filtering Ash Variables
# Threshold values for ash are: Thresh.Ash.min and Thresh.Ash.max.
# Update pipe to filter out any out-of-range Ash values. Add the additional filter at the end of the pipe.
Lab_Ash_Calculated <- Lab_Ash_Calculated %>% filter(Ash > Thresh.Ash.min & Ash < Thresh.Ash.max)
#> Error in eval(lhs, parent, parent): object 'Lab_Ash_Calculated' not found
# Summarising Ash Variables
# Use a PIPE with the grouped_by() and summarize() functions to produce a data table called Lab_Ash
# Grouped by LabID and summarises variable Ash by taking its grouped mean values
# Resulting table, Lab_Ash, must then have two variables, LabID and Ash, where LabID now contains unique values (no replicates)
Lab_Ash<-Lab_Ash_Calculated %>% grouped_by(LabID) %>% summarize(Ash=mean(Ash))
#> Error in eval(lhs, parent, parent): object 'Lab_Ash_Calculated' not found
# Import the Pol and Brix data into a data table called “Lab_PB_Data”
Lab_PB_Data <- read.table("Lab_Pol_Brix.csv", header=TRUE, sep=",", dec =".")
#> Warning in file(file, "rt"): cannot open file 'Lab_Pol_Brix.csv': No such
#> file or directory
#> Error in file(file, "rt"): cannot open the connection
# Use the mutate() function to add to the data table Lab_PB_Data a new variable, PredBrix, which uses the ExpectedBrix function with Pol as input
Lab_PB_Data <-mutate(Lab_PB_Data,PredBrix=ExpectedBrix(Pol))
#> Error in mutate(Lab_PB_Data, PredBrix = ExpectedBrix(Pol)): object 'Lab_PB_Data' not found
Created on 2019-06-01 by the reprex package (v0.3.0)
However, these files do exist in the environment:
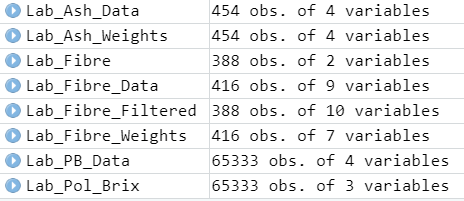
Can someone please explain why I am getting the error?