Hello!
I am an rookie in R and trying to QC the dataset from GEO.
I ran the following code :
rm(list=ls())
options(BioC_mirror="http://mirrors.tuna.tsinghua.edu.cn/bioconductor/")
options("repos" = c(CRAN="The Comprehensive R Archive Network"
))
workdir = "D:/GSEfibrosis/GSE222070"
if( !file.exists(workdir)){
if( !dir.create(workdir, showWarnings = FALSE, recursive = TRUE) ){
stop(paste("dir.create failed: workdir=",workdir,sep=""))
}
}
setwd(workdir)
getwd()
gset <- getGEO("GSE222070",getGPL = FALSE)
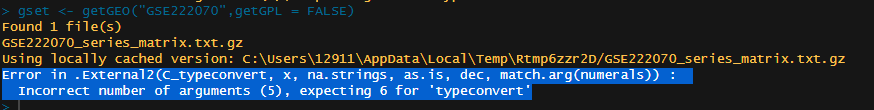
The error appeared.
gset <- getGEO("GSE222070",getGPL = FALSE)
Found 1 file(s)
GSE222070_series_matrix.txt.gz
Using locally cached version: C:\Users\12911\AppData\Local\Temp\Rtmp6zzr2D/GSE222070_series_matrix.txt.gz
Error in .External2(C_typeconvert, x, na.strings, as.is, dec, match.arg(numerals)) :
Incorrect number of arguments (5), expecting 6 for 'typeconvert'
what's problem? Can anyone help me?