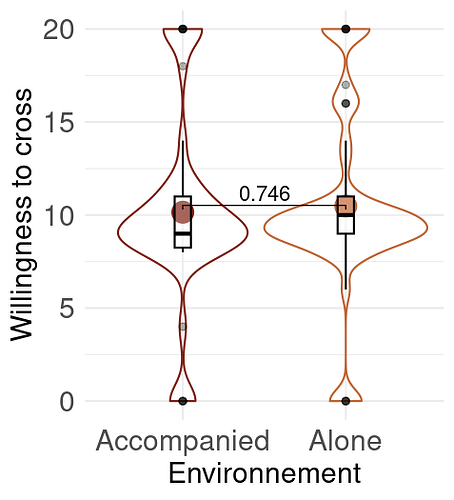
Thank you for your answer, I use these packages: ggplot2, rstatix, tydiverse, dplyr, ggpubr, MetBrewer
I'm not sure they are all usefull for this part.
Here are my data
structure(list(id = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L), levels = c("1",
"2", "3", "4", "5", "6", "7", "8", "9", "10", "11", "12", "13",
"14", "15", "16", "17", "18", "19", "20", "21", "22", "23", "24",
"25", "26", "27", "28", "29", "30", "31", "32", "33", "34", "35",
"36", "37", "38", "39", "40", "41", "42", "43", "44", "45", "46",
"47", "48", "49", "50", "51", "52", "53", "54", "55", "56", "57",
"58", "59", "60", "61", "62", "63", "64", "65", "66", "67", "68",
"69", "70", "71", "72", "73", "74", "75", "76", "77", "78", "79",
"80", "81", "82", "83", "84"), class = "factor"), duree = c(118,
118, 118, 118, 118, 118, 118, 118, 118, 118, 118, 118, 118, 118,
118, 118, 118, 118, 118, 118, 118, 118, 118, 118, 150, 150, 150,
150, 150, 150, 150, 150, 150, 150, 150, 150, 150, 150, 150, 150,
150, 150, 150, 150, 150, 150, 150, 150, 186, 186, 186, 186, 186,
186, 186, 186, 186, 186, 186, 186, 186, 186, 186, 186, 186, 186,
186, 186, 186, 186, 186, 186, 492, 492, 492, 492, 492, 492, 492,
492, 492, 492, 492, 492, 492, 492, 492, 492, 492, 492, 492, 492,
492, 492, 492, 492, 204, 204, 204, 204), gender = c("Woman",
"Woman", "Woman", "Woman", "Woman", "Woman", "Woman", "Woman",
"Woman", "Woman", "Woman", "Woman", "Woman", "Woman", "Woman",
"Woman", "Woman", "Woman", "Woman", "Woman", "Woman", "Woman",
"Woman", "Woman", "Woman", "Woman", "Woman", "Woman", "Woman",
"Woman", "Woman", "Woman", "Woman", "Woman", "Woman", "Woman",
"Woman", "Woman", "Woman", "Woman", "Woman", "Woman", "Woman",
"Woman", "Woman", "Woman", "Woman", "Woman", "Man", "Man", "Man",
"Man", "Man", "Man", "Man", "Man", "Man", "Man", "Man", "Man",
"Man", "Man", "Man", "Man", "Man", "Man", "Man", "Man", "Man",
"Man", "Man", "Man", "Woman", "Woman", "Woman", "Woman", "Woman",
"Woman", "Woman", "Woman", "Woman", "Woman", "Woman", "Woman",
"Woman", "Woman", "Woman", "Woman", "Woman", "Woman", "Woman",
"Woman", "Woman", "Woman", "Woman", "Woman", "Woman", "Woman",
"Woman", "Woman"), age = c(22, 22, 22, 22, 22, 22, 22, 22, 22,
22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 21,
21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21,
21, 21, 21, 21, 21, 21, 21, 45, 45, 45, 45, 45, 45, 45, 45, 45,
45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 21,
21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21,
21, 21, 21, 21, 21, 21, 21, 24, 24, 24, 24), eHMI = structure(c(2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L), levels = c("Absent", "Can cross", "Can not cross"
), class = "factor"), environment = structure(c(2L, 2L, 2L, 2L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L
), levels = c("Accompanied", "Alone"), class = "factor"), distance = structure(c(1L,
1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L,
1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L,
1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L,
1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L,
1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L,
1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L,
1L, 2L, 2L), levels = c("10m", "30m"), class = "factor"), speed = structure(c(2L,
1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L,
1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L,
1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L,
1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L,
1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L,
1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L,
1L, 2L, 1L), levels = c("15km/h", "7km/h"), class = "factor"),
willingness = c(10, 10, 10, 10, 11, 11, 11, NA, 11, 11, 10,
10, 11, 10, 10, 14, 10, 13, 10, 10, 10, 12, 10, 12, 9, 10,
9, 9, 8, 8, 8, 8, 8, 9, 10, 8, 9, 9, 9, 8, 10, 9, 9, 9, 10,
10, NA, 10, 10, 9, 8, 9, 9, 9, 9, 9, 9, 8, 9, 9, 9, 8, 8,
9, 8, 9, 9, 9, 9, 9, 9, 9, 20, 20, 20, 20, 20, 20, 20, 20,
0, 0, 0, 0, 0, 0, 0, 0, 14, 6, 20, 20, 18, 4, 20, 20, 17,
16, 16, 16), cluster = structure(c(3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L), levels = c("1", "2", "3", "4"), class = "factor"),
all = c("Can cross-Alone-10m-7km/h", "Can cross-Alone-10m-15km/h",
"Can cross-Alone-30m-7km/h", "Can cross-Alone-30m-15km/h",
"Can cross-Accompanied-10m-7km/h", "Can cross-Accompanied-10m-15km/h",
"Can cross-Accompanied-30m-7km/h", "Can cross-Accompanied-10m-15km/h",
"Can not cross-Alone-10m-7km/h", "Can not cross-Alone-10m-7km/h",
"Can not cross-Alone-30m-15km/h", "Can not cross-Alone-30m-15km/h",
"Can not cross-Accompanied-10m-7km/h", "Can not cross-Accompanied-10m-15km/h",
"Can not cross-Accompanied-30m-7km/h", "Can not cross-Accompanied-30m-15km/h",
"Absent-Alone-10m-7km/h", "Absent-Alone-10m-15km/h", "Absent-Alone-30m-7km/h",
"Absent-Alone-30m-15km/h", "Absent-Accompanied-10m-7km/h",
"Absent-Accompanied-10m-15km/h", "Absent-Accompanied-30m-7km/h",
"Absent-Accompanied-30m-15km/h", "Can cross-Alone-10m-7km/h",
"Can cross-Alone-10m-15km/h", "Can cross-Alone-30m-7km/h",
"Can cross-Alone-30m-15km/h", "Can cross-Accompanied-10m-7km/h",
"Can cross-Accompanied-10m-15km/h", "Can cross-Accompanied-30m-7km/h",
"Can cross-Accompanied-10m-15km/h", "Can not cross-Alone-10m-7km/h",
"Can not cross-Alone-10m-7km/h", "Can not cross-Alone-30m-15km/h",
"Can not cross-Alone-30m-15km/h", "Can not cross-Accompanied-10m-7km/h",
"Can not cross-Accompanied-10m-15km/h", "Can not cross-Accompanied-30m-7km/h",
"Can not cross-Accompanied-30m-15km/h", "Absent-Alone-10m-7km/h",
"Absent-Alone-10m-15km/h", "Absent-Alone-30m-7km/h", "Absent-Alone-30m-15km/h",
"Absent-Accompanied-10m-7km/h", "Absent-Accompanied-10m-15km/h",
"Absent-Accompanied-30m-7km/h", "Absent-Accompanied-30m-15km/h",
"Can cross-Alone-10m-7km/h", "Can cross-Alone-10m-15km/h",
"Can cross-Alone-30m-7km/h", "Can cross-Alone-30m-15km/h",
"Can cross-Accompanied-10m-7km/h", "Can cross-Accompanied-10m-15km/h",
"Can cross-Accompanied-30m-7km/h", "Can cross-Accompanied-10m-15km/h",
"Can not cross-Alone-10m-7km/h", "Can not cross-Alone-10m-7km/h",
"Can not cross-Alone-30m-15km/h", "Can not cross-Alone-30m-15km/h",
"Can not cross-Accompanied-10m-7km/h", "Can not cross-Accompanied-10m-15km/h",
"Can not cross-Accompanied-30m-7km/h", "Can not cross-Accompanied-30m-15km/h",
"Absent-Alone-10m-7km/h", "Absent-Alone-10m-15km/h", "Absent-Alone-30m-7km/h",
"Absent-Alone-30m-15km/h", "Absent-Accompanied-10m-7km/h",
"Absent-Accompanied-10m-15km/h", "Absent-Accompanied-30m-7km/h",
"Absent-Accompanied-30m-15km/h", "Can cross-Alone-10m-7km/h",
"Can cross-Alone-10m-15km/h", "Can cross-Alone-30m-7km/h",
"Can cross-Alone-30m-15km/h", "Can cross-Accompanied-10m-7km/h",
"Can cross-Accompanied-10m-15km/h", "Can cross-Accompanied-30m-7km/h",
"Can cross-Accompanied-10m-15km/h", "Can not cross-Alone-10m-7km/h",
"Can not cross-Alone-10m-7km/h", "Can not cross-Alone-30m-15km/h",
"Can not cross-Alone-30m-15km/h", "Can not cross-Accompanied-10m-7km/h",
"Can not cross-Accompanied-10m-15km/h", "Can not cross-Accompanied-30m-7km/h",
"Can not cross-Accompanied-30m-15km/h", "Absent-Alone-10m-7km/h",
"Absent-Alone-10m-15km/h", "Absent-Alone-30m-7km/h", "Absent-Alone-30m-15km/h",
"Absent-Accompanied-10m-7km/h", "Absent-Accompanied-10m-15km/h",
"Absent-Accompanied-30m-7km/h", "Absent-Accompanied-30m-15km/h",
"Can cross-Alone-10m-7km/h", "Can cross-Alone-10m-15km/h",
"Can cross-Alone-30m-7km/h", "Can cross-Alone-30m-15km/h"
), allcode = structure(c(1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L,
9L, 10L, 11L, 12L, 13L, 14L, 15L, 16L, 17L, 18L, 19L, 20L,
21L, 22L, 23L, 24L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
11L, 12L, 13L, 14L, 15L, 16L, 17L, 18L, 19L, 20L, 21L, 22L,
23L, 24L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L,
13L, 14L, 15L, 16L, 17L, 18L, 19L, 20L, 21L, 22L, 23L, 24L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 13L, 14L,
15L, 16L, 17L, 18L, 19L, 20L, 21L, 22L, 23L, 24L, 1L, 2L,
3L, 4L), levels = c("0000", "0001", "0010", "0011", "0100",
"0101", "0110", "0111", "1000", "1001", "1010", "1011", "1100",
"1101", "1110", "1111", "2000", "2001", "2010", "2011", "2100",
"2101", "2110", "2111"), class = "factor"), ido = c(1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63,
64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78,
79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93,
94, 95, 96, 97, 98, 99, 100)), row.names = c(NA, -100L), class = c("tbl_df",
"tbl", "data.frame"))
![]() Error occurred in the 4th layer.
Error occurred in the 4th layer.![]() The following aesthetics are invalid:
The following aesthetics are invalid:![]()
![]() Did you mistype the name of a data column or forget to add
Did you mistype the name of a data column or forget to add