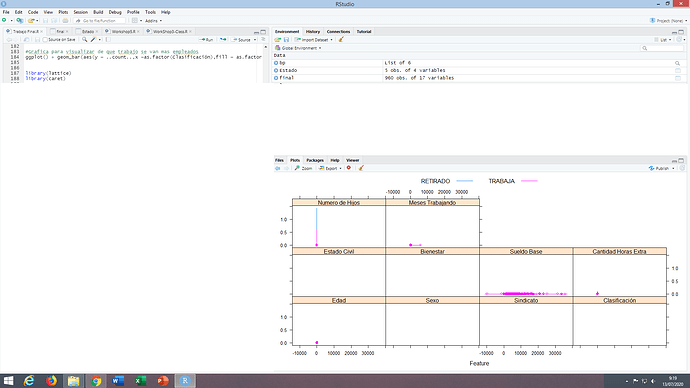
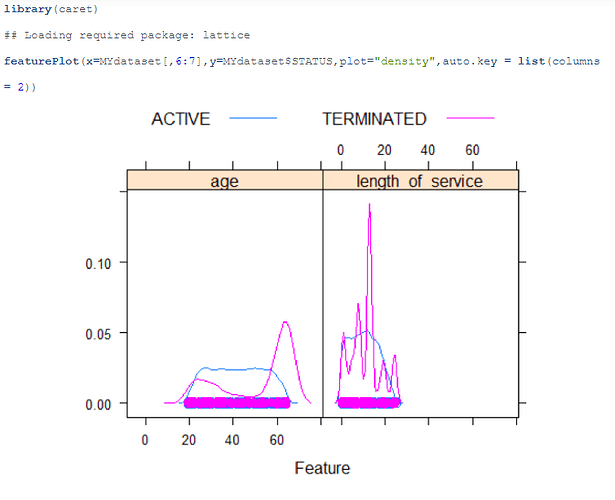
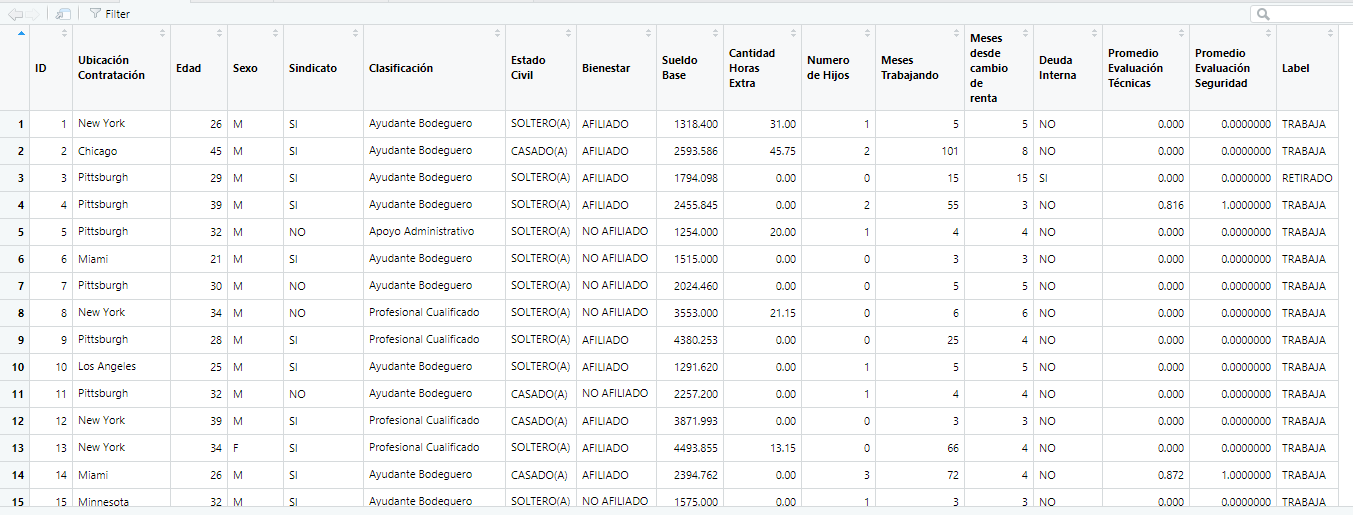
I tried changing the Label data type to factor and i reicive this plot:
and i have this Warning messages:
1: In (function (x, darg, groups = NULL, weights = NULL, subscripts = TRUE, :
NAs introducidos por coerción
2: In (function (x, darg, groups = NULL, weights = NULL, subscripts = TRUE, :
NAs introducidos por coerción
3: In (function (x, darg, groups = NULL, weights = NULL, subscripts = TRUE, :
NAs introducidos por coerción
4: In (function (x, darg, groups = NULL, weights = NULL, subscripts = TRUE, :
NAs introducidos por coerción
5: In (function (x, darg, groups = NULL, weights = NULL, subscripts = TRUE, :
NAs introducidos por coerción
6: In panel.superpose(x, darg = darg, plot.points = plot.points, ref = FALSE, :
NAs introducidos por coerción
7: In panel.superpose(x, darg = darg, plot.points = plot.points, ref = FALSE, :
NAs introducidos por coerción
8: In panel.superpose(x, darg = darg, plot.points = plot.points, ref = FALSE, :
NAs introducidos por coerción
9: In panel.superpose(x, darg = darg, plot.points = plot.points, ref = FALSE, :
NAs introducidos por coerción
10: In panel.superpose(x, darg = darg, plot.points = plot.points, ref = FALSE, :
NAs introducidos por coerción
So i apply this code for ommite NA :
final<-na.omit(final)
And again the plot gives me as a result : NULL