Hi,
I have the data, which can be found Dropbox
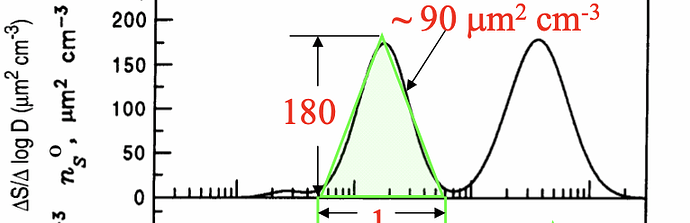
I need to plot something like
So, the data header is given 15.1 to 661 which is the x-axis and the rest are the y-axis points.
So, I want to make a distribution curve like the above figure.
Could you please let me know how to make it?
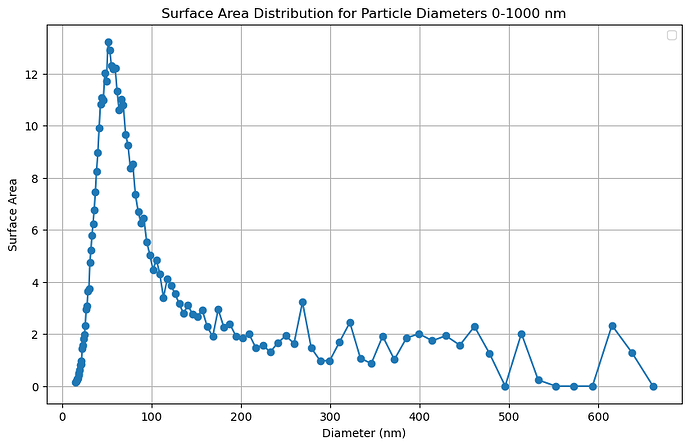
I tried with julius.ai and from that I got this kind of plot
import pandas as pd
import matplotlib.pyplot as plt
# Load the data with corrected headers
headers = [15.1, 15.7, 16.3, 16.8, 17.5, 18.1, 18.8, 19.5, 20.2, 20.9, 21.7, 22.5, 23.3, 24.1, 25, 25.9, 26.9, 27.9, 28.9, 30, 31.1, 32.2, 33.4, 34.6, 35.9, 37.2, 38.5, 40, 41.4, 42.9, 44.5, 46.1, 47.8, 49.6, 51.4, 53.3, 55.2, 57.3, 59.4, 61.5, 63.8, 66.1, 68.5, 71, 73.7, 76.4, 79.1, 82, 85.1, 88.2, 91.4, 94.7, 98.2, 101.8, 105.5, 109.4, 113.4, 117.6, 121.9, 126.3, 131, 135.8, 140.7, 145.9, 151.2, 156.8, 162.5, 168.5, 174.7, 181.1, 187.7, 194.6, 201.7, 209.1, 216.7, 224.7, 232.9, 241.4, 250.3, 259.5, 269, 278.8, 289, 299.6, 310.6, 322, 333.8, 346, 358.7, 371.8, 385.4, 399.5, 414.2, 429.4, 445.1, 461.4, 478.3, 495.8, 514, 532.8, 552.3, 572.5, 593.5, 615.3, 637.8, 661.2]
df = pd.read_csv('DATA_Sample_SurArea_Dist.csv', names=headers, skiprows=1, encoding='UTF-8-SIG')
# Filter the data for particle diameters from 0 to 1000 nm
filtered_df = df.loc[:, df.columns <= 1000]
# Plotting
plt.figure(figsize=(10, 6), facecolor='white')
plt.plot(filtered_df.columns, filtered_df.iloc[0], marker='o', linestyle='-')
plt.title('Surface Area Distribution for Particle Diameters 0-1000 nm')
plt.xlabel('Diameter (nm)')
plt.ylabel('Surface Area')
plt.grid(True)
plt.show()
I want to make it something like a plot but with R.
please help out.
Thanks.