Hi,
I would like to create a Shiny app that allows user to upload a csv and image files and draw the image file by placing rectangles according to the pixel coordinate data in the uploaded csv file. The file uploading part is working. I am trying to get the image drawing part working with Shiny using the following functions from the magick package:
- image <- magick::image_read(req(input$imported$datapath)), which reads the uploaded image (inputID="imported") as a magick-image object. This is working.
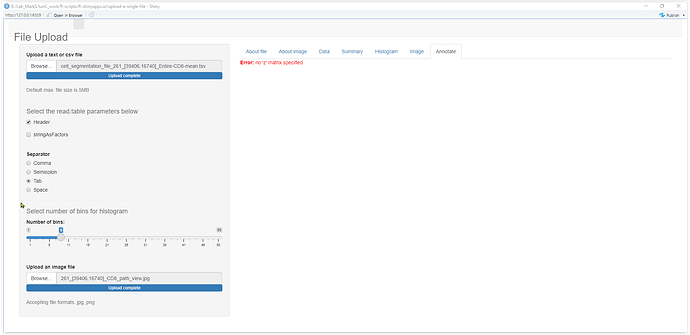
- magick::image_draw(image=image()), which allows me to draw the imported image in a local R script But here it has returned an error "Warning: Error in image.default: no 'z' matrix specified."
- graphics::rect(), which places rectangles on the image according to the four position arguments xleft, xright, ytop, and ybottom. This is working in my local R script. I have commented this part out in the provided server.R file.
- grDevices::dev.off(), which closes the image device in my local R script. This part is commented out in the server.R
I am looking to do the file uploading and image annotation under different tabPanels listed at the end of the server.R. Any thoughts on making the image_draw() function working with Shiny? Here is a printscreen of the tabPanel that returns the error
My ui.R
library(shiny)
library(magick)
# Define User Interface
shinyUI(fluidPage(
navbarPage(
# App title ----
titlePanel("File Upload"),
# Sidebar layout with input and output definitions ----
sidebarLayout(
sidebarPanel(
# fileinput() function is used to get the file upload control option
fileInput(inputId = "file", label = "Upload a text or csv file"),
helpText("Default max. file size is 5MB"),
# Add a horizontal line
tags$hr(),
# Add a level 4 header
h4(helpText("Select the read.table parameters below")),
checkboxInput(inputId = 'header', label = 'Header', value = FALSE),
checkboxInput(inputId = "stringAsFactors", "stringAsFactors", FALSE),
br(),
radioButtons( inputId = 'sep'
,label = 'Separator'
,choices = c(Comma=',',Semicolon=';',Tab='\t', Space='')
,selected = ','),
# Add a horizontal line
tags$hr(),
h4(helpText("Select number of bins for histogram")),
# Input: Slider for the number of bins ----
sliderInput(inputId = "bins",
label = "Number of bins:",
min = 1,
max = 50,
value = 30),
# Add a horizontal line
tags$hr(),
fileInput(inputId = "imported", label = "Upload an image file"),
helpText("Accepting file formats: jpg, png")
),
mainPanel(
uiOutput("tb")
)
)
))
)
My server.R
shinyServer(function(input,output){
# This reactive function will take the inputs from ui.R and use them for read.table() to read the data from the file. It returns the dataset in the form of a data.frame.
# To use the input, use input$inputID (e.g., input$file)
# To use the data data.frame, refer it as "data()" rather than "data"
data <- reactive({
file1 <- input$file
if(is.null(file1)){return()}
# file$datapath -> gives the path of the file
read.table( file=file1$datapath
,sep=input$sep
,header = input$header
,stringsAsFactors = input$stringAsFactors)
})
# This reactive output contains the summary of the dataset and display the summary in table format
output$file_1 <- renderTable({
if(is.null(data())){return ()}
input$file
})
# This reactive output contains the summary of the dataset and display the summary in table format
output$file_2 <- renderTable(
input$imported
)
# This reactive output contains the summary of the dataset and display the summary in table format
output$sum <- renderTable({
if(is.null(data())){return ()}
summary(data())
})
# This reactive output contains the dataset and display the dataset in table format
output$table <- renderTable({
if(is.null(data())){return ()}
data()
})
# Make a histogram on column 3 of input data file
output$distPlot <- renderPlot({
# Get column 3
x <- data()[,3]
x <- na.omit(x)
bins <- seq(min(x), max(x), length.out = input$bins + 1)
hist(x
,breaks = bins
, col = "#75AADB"
, border = "black"
,xlab = "Marker measurement"
,main = "Histogram of marker measurement"
,density=TRUE )
})
# Display the uploaded image
output$image <- renderImage({
# The req function makes sure a input was actually selected
req(input$imported)
list(
src = normalizePath(file.path(input$imported$datapath))
,alt = "there should be an image here"
#,width = 400
#,height = 400
)
}, deleteFile = FALSE)
#--------------------------------------------------
# Edit the uploaded image file using magick package
#--------------------------------------------------
loaded_image <- reactive({
# Create pixel coordinates for rectangles
rect_width_half <- 12.5
rect_x_left <- data()[,1] - rect_width_half
rect_x_right <- data()[,1] + rect_width_half
rect_y_top <- data()[,2] - rect_width_half
rect_y_bottom <- data()[,2] + rect_width_half
image <- magick::image_read(req(input$imported$datapath))
magick::image_draw(image=image())
#return(image_drew)
# image <- magick::image_read(req(input$imported$datapath))
# magick::image_draw(image())
# ## Draw rectangles for individual stained cells
# graphics::rect( xleft= rect_x_left
# ,ybottom=rect_y_bottom
# ,xright=rect_x_right
# ,ytop=rect_y_top
# ,col = NA # color(s) to fill or shade the rectangle(s) with. The default NA (or also NULL) means do not fill, i.e., draw transparent rectangles, unless density is specified.
# ,border = "black" # color for rectangle border(s).
# ,lty = par("lty")
# ,lwd = 2)
#grDevices::dev.off()
})
output$image_edited <- renderPlot({
image_ggplot(loaded_image())
})
# The following renderUI is used to dynamically generate the tabsets when the file is loaded. Until the file is loaded, app will not show the tabset.
output$tb <- renderUI({
if(is.null(data()))
h5("Powered by", tags$img(src='RStudio-Ball.png', heigth=200, width=200))
else
tabsetPanel( tabPanel("About file", tableOutput(outputId ="file_1"))
,tabPanel("About image", tableOutput(outputId = "file_2"))
,tabPanel("Data", tableOutput(outputId ="table"))
,tabPanel("Summary", tableOutput(outputId ="sum"))
,tabPanel("Histogram",plotOutput(outputId = "distPlot"))
,tabPanel("Image", imageOutput(outputId = "image"))
,tabPanel("Annotate", imageOutput(outputId = "image_edited"))
)
})
})
Any feedback would be highly appreciated.
Chang