Double exponential model is very useful for modeling decomposition for surface and incorporated litter. Please I don't know if any expert in modeling this nonlinear decomposition using the nls, nlme, or nlraa package can help me out. I have added a fake dataset and an image of the double exponential decomposition model below. I am very greatful if someone can help me or point me to where I can get solution for this. I need the model and also I want to be able to plot the coeffients from this model in ggplot2, but if I can get the code to fit this nonlinear model in R using nlme, I can figure out the plotting my self
Load packages
require(lmerTest)
require(nlme)
require(ggplot2)
require(cowplot)
require(lmtest)
Single Exponential Decomposition Code
Data
decomp <- data.frame(
Weight_loss = c(55, 40, 30, 20, 24, 18),
time = c(10, 14, 28, 30, 55, 40),
Placement = c(
'surface',
'incorporated',
'surface',
'incorporated',
'surface',
'incorporated'
)
)
Double exponential decomposition model
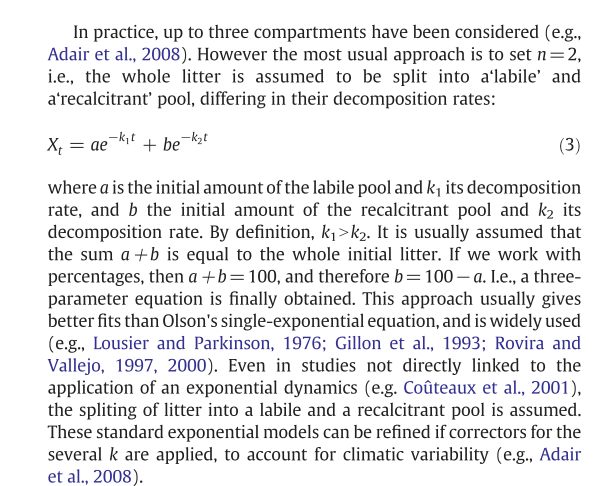
#Yang & Janssen model (# Single exponential decomposition model)
decomp2=nlme(Frac~exp(-k*(Time^a)),
data=dato,
fixed=k+a~1,
random=k+a~1,
groups=~Pod,
method='REML',
control=ctrl4,
start=c(1,0.5)) #Beware of local optimum, using a=0.3 gives similar estimates but higher Rsquare (0.91)
summary(decomp2)
RMSE_decomp2=sqrt(sum(residuals(decomp2)^2)/nrow(as.data.frame(decomp2$residuals)))
R_yan=summary(lm(predict(decomp2)~dato$Frac))$r.squared
intervals(decomp1,which='fixed') # for 95% CI Olson model
intervals(decomp2,which='fixed') # for 95% CI Yang & Janssen model
RMSE_decomp1
RMSE_decomp2
R_ols
R_yan
#prediction of C loss at days 14, 30, 365
t14=data.frame(Time=c(0,14,30, 365))
1-predict(decomp2,newdata=t14,level=0)
Time=seq(0,70,by=0.5) # replace 70 with 365 if one wants to predict over 1 year
Frac=exp(-fixef(decomp2)[1]*(Time^fixef(decomp2)[2]))
gag1=data.frame(cbind(Time,Frac))
P04=ggplot(dato,aes(x=Time,y=100Frac))+
geom_point(aes(shape=paste(Batch,Pod)),
size=2.5)+
geom_line(data=gag1,aes(x=Time,100Frac),
colour='black',
size=1,
linetype=1)+
xlab('Time (days)')+
ylab('Residual DM weight (%)')+
#ggtitle('CPH decay at 20deg C 100% RH')+
theme(legend.position='none')