In a nutshell, I need to be able to run a document term matrix from a Twitter dataset within an XGBoost classifier. I have completed the document term matrix, but I am missing some key part of preparing the DTM and putting it in a format that the model will accept. I know that you have to convert the DYM back to a data frame, and then you have to create the "training" and "testing" partitions. Can someone put me on the right track as far as the code that I am missing?
Here is the code for the Natural Language Processing part:
setwd('C:/rscripts/random_forest')
dataset = read.csv('tweets_all.csv', stringsAsFactors = FALSE)
library(tm)
corpus <- iconv(dataset$text, to = "utf-8")
corpus <- Corpus(VectorSource(corpus))
inspect(corpus[1:5])
corpus <- tm_map(corpus, tolower)
corpus <- tm_map(corpus, removePunctuation)
corpus <- tm_map(corpus, removeNumbers)
cleanset <- tm_map(corpus, removeWords, stopwords('english'))
#my_custom_stopwords <- c("ââ¬\u009dpotus", "Ã\u009dÃ", "Ã\u009djoebiden", "Ã\u009dand", "Ã\u009dhillary", "ââ¬Å¾Ã")
#cleanset <- tm_map(corpus, removeWords, my_custom_stopwords)
removeURL <- function(x) gsub('http[[:alnum:]]*', '', x)
cleanset <- tm_map(cleanset, content_transformer(removeURL))
cleanset <- tm_map(cleanset, stripWhitespace)
cleanset <- tm_map(cleanset, removeWords, c('Ã\u009dhillary','ââ¬Å¾Ã','ââ¬Å¡Ã','just','are','all','they'))
tdm <- TermDocumentMatrix(cleanset)
tdm <- as.matrix(tdm)
Here is the code that I found to use for the XGBoost classification model. It is currently written to accommodate a different dataset (i.e. mushroom data), but I was going to recycle this code to use with my document term matrix from my text mining.
library(caret)
library(xgboost)
install.packages('e1071', dependencies=TRUE)
mushroom_data$cap.shape = as.factor(mushroom_data$cap.shape)
newvars <- dummyVars( ~ cap.shape + cap.surface + cap.color + bruises + odor + gill.attachment + gill.spacing +
gill.size + gill.color + stalk.shape + stalk.root + stalk.surface.above.ring +
stalk.surface.below.ring + stalk.color.above.ring + stalk.color.below.ring+
veil.color + ring.number + ring.type + spore.print.color+ population+
habitat ,data=mushroom_data)
newvars <- predict(newvars, mushroom_data)
cv.ctrl <- trainControl(method = "repeatedcv", repeats = 1,number = 4,allowParallel=T)
xgb.grid <- expand.grid(nrounds = 40,eta = c(0.5,1),max_depth = c(7,10),gamma = c(0,0.2),colsample_bytree=c(1),min_child_weight=c(0.1,0.9))
xgb_tune <-train( newvars , mushroom_data$class,
method="xgbTree",
trControl=cv.ctrl,
tuneGrid=xgb.grid
)
pred = predict(xgb_tune,newvars)
mushroom_data$labels[mushroom_data$class=="e"] = 1
mushroom_data$labels[mushroom_data$class=="p"] = 0
mtrain <- xgb.DMatrix(data = newvars , label = as.matrix(mushroom_data$labels))
result_model <- xgboost(data = mtrain ,max_depth = 7, eta = 1, nthread = 4, nrounds = 40, objective = "reg:logistic", verbose = 1)
pred <- predict(result_model, mtrain)
pred[pred<0.5] = 0
pred[pred>0.5] = 1
joined = data.frame(mushroom_data$labels,pred)
joined_diff = joined$mushroom_data.labels- joined$pred
sum(joined_diff)
Can someone please show me how to place my DTM appropriately into the XGBoost code? I can provide the dataset as well.
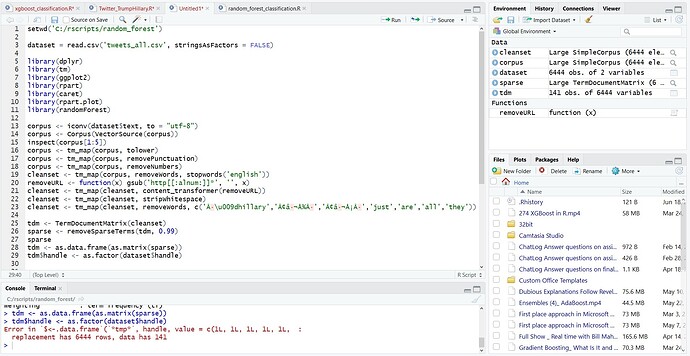

 ) and also paste in the error message (it usually helps to format these as code too, since they are often written assuming they will be displayed with a fixed-width font).
) and also paste in the error message (it usually helps to format these as code too, since they are often written assuming they will be displayed with a fixed-width font).