Dimension Reduction with UMAP and t-SNE
Authors: Dean Smith
Working with Shiny more than 1 year
Abstract: This shiny app can be used to perform dimension reduction with UMAP and t-SNE on an input file or R library dataset.
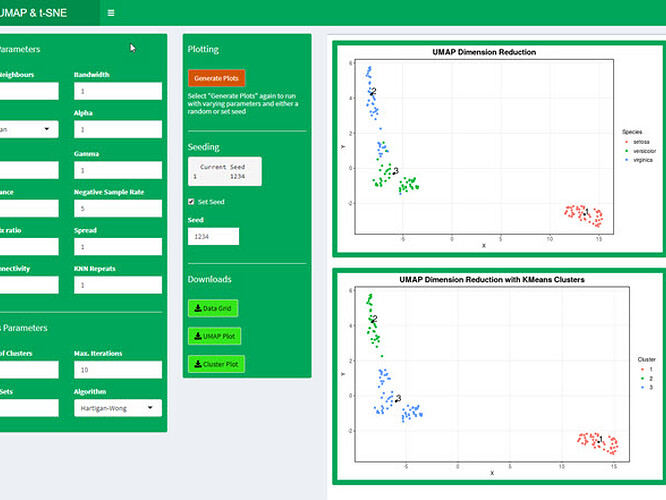
Full Description: UMAP and t-SNE are two popular non-linear dimension reduction algorithms. This shiny app will take data from either a user file input or a selection of datasets from the MASS and ggplot2 libraries.
Data pre-processing includes reducing observations, removing missing values, imputing missing values, applying a function, scaling and rounding. Any number of numeric dimensions can be selected for dimension reduction and a class can be added to the data for visualization.
UMAP and t-SNE algorithms can be run any number of times with varying parameters and either random or set seeds.
KMeans clusters are generated and parameters can be varied. The clusterID’s are added to the input data which can be downloaded and used in classification models.
Plots can be customized and downloaded.
Possible reasons why charts are not working:
-
Selected dimensions are not numeric.
-
The selected dimensions contain a dimension that looks numeric but is not, an example is the diamonds dataset, the dimension “table” is not numeric – it can be used as a class.
-
Dataset contains missing values, an example is the biopsy dataset, see if you can work out which dimension contains missing values – these can be removed in processing.
Enjoy!
Category: Research
Keywords: dimension reduction, KMeans, UMAP, t-SNE
Shiny app: https://atamaianalytics.shinyapps.io/DimRedWithUMAPandtSNE/
Repo: GitHub - atamaianalytics/DimensionReduction: Shiny app for dimension reduction using UMAP and t-SNE
RStudio Cloud: Posit Cloud
Thumbnail:
Full image: