Hello all,
I reached out to the R-devel community for a question, but it didn't get accepted. So I will try again here. It is concerning differences in matrix multiplication for large matrices using different blas libraries on different environments.
As I am representing a rather large user community at BASF and this is critical for our day to day work, I would really appreciate any light that could be shed on this topic.
With kind regards,
r-devel-owner@r-project.org|9:40 AM (18 minutes ago)
to me
The message's content type was not explicitly allowed
---------- Forwarded message ----------
From: ... <[...)>
To: r-devel@r-project.org
Cc:
Bcc:
Date: Fri, 29 Mar 2019 15:15:51 +0100
Subject: Difference in results while performing matrix multiplication
Hello R-devel,
While performing some migration from cluster A to B, one of my users has noticed something strange in the matrix multiplication.
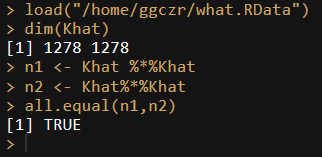
The old environment:
sessionInfo()
R version 3.4.1 (2017-06-30)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Red Hat Enterprise Linux Server release 6.9 (Santiago)
Matrix products: default
BLAS: /gpfs/gssgpfs1/biogrid/tools/bioinfo/app/R-3.4.1/lib64/R/lib/libRblas.so
LAPACK: /gpfs/gssgpfs1/biogrid/tools/bioinfo/app/R-3.4.1/lib64/R/lib/libRlapack.so
the new environment
sessionInfo()
R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Red Hat Enterprise Linux Server 7.4 (Maipo)
Matrix products: default
BLAS/LAPACK: /gpfs/gssgpfs1/biogrid/tools/eb/software/OpenBLAS/0.3.1-GCC-7.3.0-2.30/lib/[libopenblas_haswellp-r0.3.1.so](http://libopenblas_haswellp-r0.3.1.so/)
This is clearly the difference: BLAS/LAPACK
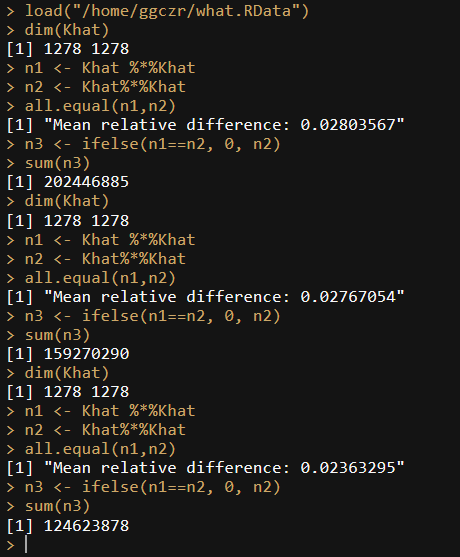
When performed on the old environment we get the following:
On the new environment we get this
To make matters even worse, the result changes at each run, so this can't be correct right?
Could anybody shed some light here on what might be going on?
with kind regards,