I am sorry that I have posted this issue in two places, I know that this isn't proper practice, but it is not allowing me to edit my previous post.
I am trying to determine the slope of two GAM models and compare how the slope changes between models.
I created GAM models looking at the relationship between precipitation and stream discharge under a pre-and post-treatment scenario. The pre-treatment model is named "preGAM" and the post-treatment model is named "postGAM". The code used to create these models is provided below.
I first created both GAM models using the following code:
preGAM <- gam(preGAM.dat$DailyIncrease ~ s(preGAM.dat$Precipitation, method = "REML"))
postGAM <- gam(postGAM.dat$DailyIncrease ~ s(postGAM.dat$Precipitation, method = "REML"))
I then viewed the summary of the model "preGAM", which returned the following output:
> summary(preGAM)
Family: gaussian
Link function: identity
Formula:
preOutlierRem.dat$DailyIncrease ~ s(preOutlierRem.dat$Precipitation)
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.1275 0.0186 6.853 1.97e-11 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(preOutlierRem.dat$Precipitation) 6.102 7.195 40.2 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
R-sq.(adj) = 0.347 Deviance explained = 35.4%
-REML = 335.04 Scale est. = 0.18963 n = 548```
I then viewed the summary for the model "postGAM", which returned the following output:
Family: gaussian
Link function: identity
Formula:
postOutlierRem.dat$DailyIncrease ~ s(postOutlierRem.dat$Precipitation)
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.13046 0.01273 10.25 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(postOutlierRem.dat$Precipitation) 8.836 8.991 101.2 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
R-sq.(adj) = 0.452 Deviance explained = 45.6%
-REML = 636.78 Scale est. = 0.17773 n = 1097
I then plotted each relationship as shown in Figures 1 and 2 below:
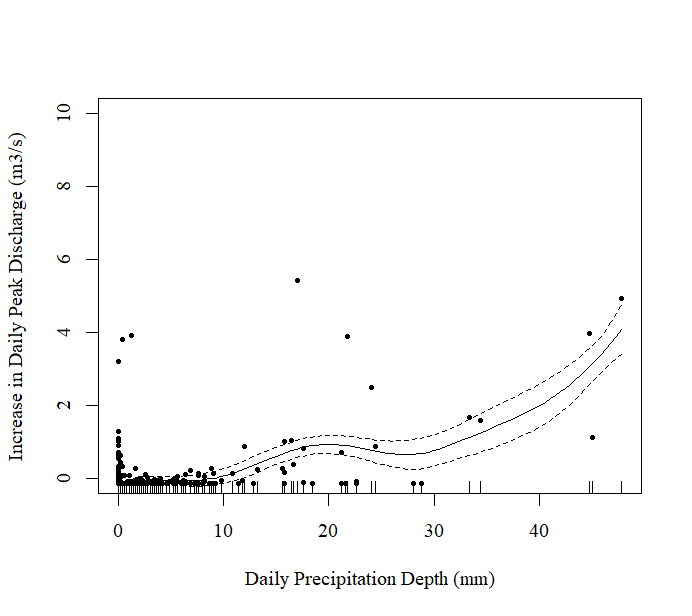
Figure 1: Relationship Between Daily Precipitation Depth (mm) and the Increase in Daily Peak Discharge (m3/s) Pre-Treatment
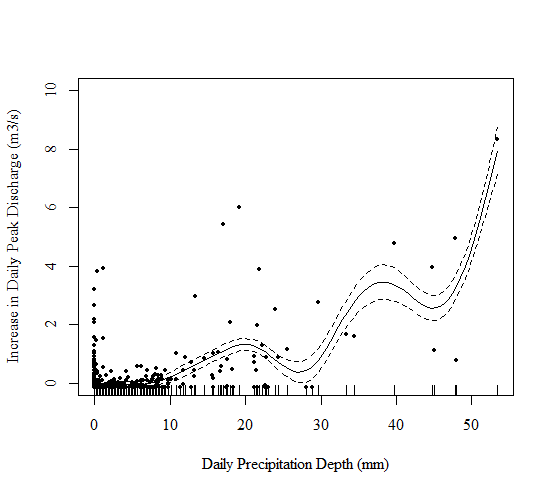
Figure 2: Relationship Between Daily Precipitation Depth (mm) and the Increase in Daily Peak Discharge (m3/s) Post-Treatment
I am now trying to make sense of these outputs and determine how to 1) understand the preGAM relationship between the variables Precipitation and Daily Increase, 2) understand the postGAM relationship between the variables Precipitation and Daily Increase, and 3) make observations as to if and how the relationship between Precipitation and Daily Increase preGAM and postGAM differ.
I began by looking at the significance of the significance of the smoothed term for precipitation in both models. As shown in the output above, the smoothed term for precipitation was highly significant for both the preGAM (p < 2e-16) and postGAM (p <2e-16) scenarios.
As I understand it, the value of the effective degrees of freedom (edf) represents the order of the relationship, where an EDF of 1 is equivalent to a linear relationship, an edf of 2 is equivalent to a quadratic curve, and so on, with higher edfs describing more "wiggly" curves.
Therefore, my preGAM model has been fitted an order 6 (edf = 6.102) relationship, and my postGAM model has been fitted to an order 9 (edf = 8.836) relationship. As the p-value for the approximate significance of the smoothed term (precipitation) for the preGAM model is <2e-16, and and the p-value for the approximate significance for the smoothed term (precipitation) for the postGAM model is <2e-16. I know that for both models, the relationship between daily precipitation depth and the increase in daily stream discharge is highly significant (which makes sense logically). I believe that for comparison these should have the same model order in order to accurately compare the slopes.
My remaining questions are:
- Does anyone have any advice on how to revise the models to ensure that both models (pre-GAM and post-GAM) are of the same order?
- Does anyone have any advice on how to determine the overall slope of the models
I want to create two models of the same model order, and compare the slopes of these models to determine if the relationship between daily precipitation depth and daily peak stream discharge is different in the post development scenario (compared to the pre-development scenario). I appreciate any advice that anyone might have.
TIA