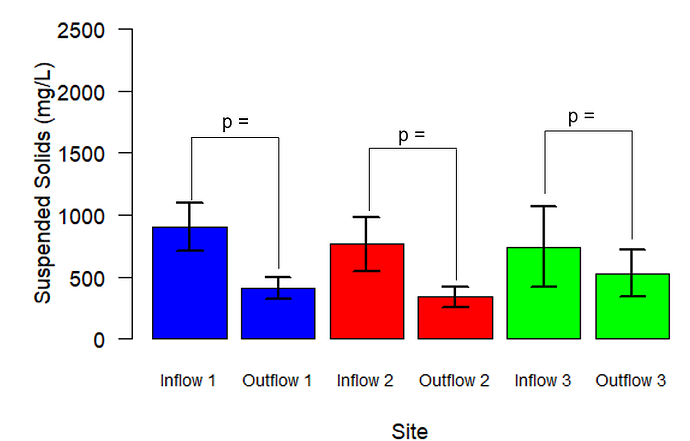
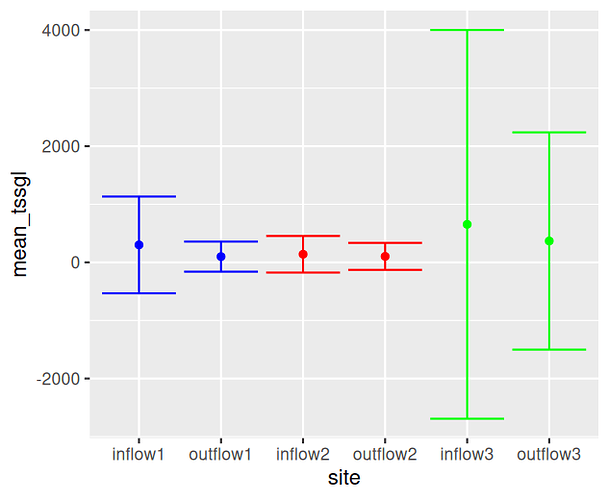
Thank you. I just posted my data below. Note that the site names in my raw data correspond to street names, and are thus different than what is shown in the graph. "Inflow 1" = "Hinton Inflow Front Cell", "Outflow 1" = "Hinton Outflow", "Inflow 2" = "Cuyler and Dewe Inflow", "Outflow 2" = "Cuyler and Dewe Outflow", "Inflow 3" = "Cuyler and Current Inflow", and "Outflow 3" = "Cuyler and Current Outflow"
dput(head(rainfall,100))
structure(list(Rainfall.Number = c(1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 7L, 7L,
7L, 7L, 7L, 7L, 7L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L,
8L, 9L, 9L, 9L, 9L, 9L, 9L, 9L, 9L, 9L, 10L, 10L, 10L, 10L, 10L,
10L, 10L, 10L, 10L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 12L, 12L,
12L), Date = c("2022-08-05", "2022-08-05", "2022-08-05", "2022-08-05",
"2022-08-05", "2022-08-05", "2022-08-05", "2022-08-05", "2022-08-05",
"2022-08-18", "2022-08-18", "2022-08-18", "2022-08-18", "2022-08-18",
"2022-08-18", "2022-08-18", "2022-08-18", "2022-08-18", "2022-08-18",
"2022-08-23", "2022-08-23", "2022-09-09", "2022-09-09", "2022-09-09",
"2022-09-09", "2022-09-09", "2022-09-09", "2022-09-09", "2022-09-09",
"2022-09-09", "2022-09-09", "2022-09-09", "2022-09-25", "2022-09-25",
"2022-09-25", "2022-09-25", "2022-09-25", "2022-09-25", "2022-09-25",
"2022-09-25", "2022-09-25", "2022-09-25", "2022-10-12", "2022-10-12",
"2022-10-12", "2022-10-12", "2022-10-12", "2022-10-12", "2022-10-12",
"2022-10-12", "2022-10-12", "2022-10-12", "2022-10-12", "2022-11-06",
"2022-11-06", "2022-11-06", "2022-11-06", "2022-11-06", "2022-11-06",
"2022-11-06", "2023-04-29", "2023-04-29", "2023-04-29", "2023-04-29",
"2023-04-29", "2023-04-29", "2023-04-29", "2023-04-29", "2023-04-29",
"2023-04-29", "2023-04-29", "2023-04-29", "2023-05-19", "2023-05-19",
"2023-05-19", "2023-05-19", "2023-05-19", "2023-05-19", "2023-05-19",
"2023-05-19", "2023-05-19", "2023-06-24", "2023-06-24", "2023-06-24",
"2023-06-24", "2023-06-24", "2023-06-24", "2023-06-24", "2023-06-24",
"2023-06-24", "2023-06-29", "2023-06-29", "2023-06-29", "2023-06-29",
"2023-06-29", "2023-06-29", "2023-06-29", "2023-07-09", "2023-07-09",
"2023-07-09"), Site = c("Hinton Upstream", "Hinton Inflow Front Cell",
"Hinton Outflow", "Hinton Downstream", "Blanchard Upstream",
"Blanchard Inflow", "Blanchard Downstream", "Cuyler and Dewe Inflow",
"Cuyler and Dewe Outflow", "Hinton Upstream", "Hinton Inflow Front Cell",
"Hinton Inflow Back Cell", "Hinton Outflow", "Hinton Downstream",
"Blanchard Upstream", "Blanchard Inflow", "Blanchard Downstream",
"Cuyler and Current Inflow", "Cuyler and Current Outflow", "Cuyler and Current Inflow",
"Cuyler and Current Outflow", "Hinton Upstream", "Hinton Inflow Front Cell",
"Hinton Outflow", "Hinton Downstream", "Blanchard Upstream",
"Blanchard Inflow", "Blanchard Downstream", "Cuyler and Dewe Inflow",
"Cuyler and Dewe Outflow", "Cuyler and Current Inflow", "Cuyler and Current Outflow",
"Hinton Upstream", "Hinton Inflow Front Cell", "Hinton Inflow Back Cell",
"Hinton Outflow", "Hinton Downstream", "Blanchard Upstream",
"Blanchard Inflow", "Blanchard Downstream", "Cuyler and Current Inflow",
"Cuyler and Current Outflow", "Hinton Upstream", "Hinton Inflow Front Cell",
"Hinton Outflow", "Hinton Downstream", "Blanchard Upstream",
"Blanchard Inflow", "Blanchard Downstream", "Cuyler and Dewe Inflow",
"Cuyler and Dewe Outflow", "Cuyler and Current Inflow", "Cuyler and Current Outflow",
"Hinton Upstream", "Hinton Inflow Front Cell", "Hinton Outflow",
"Hinton Downstream", "Blanchard Upstream", "Blanchard Inflow",
"Blanchard Downstream", "Blanchard Upstream", "Blanchard Inflow",
"Blanchard Downstream", "Hinton Upstream", "Hinton Inflow Front Cell",
"Hinton Inflow Back Cell", "Hinton Outflow", "Hinton Downstream",
"Cuyler and Dewe Inflow", "Cuyler and Dewe Outflow", "Cuyler and Current Inflow",
"Cuyler and Current Outflow", "Blanchard Upstream", "Blanchard Inflow",
"Blanchard Downstream", "Hinton Upstream", "Hinton Inflow Front Cell",
"Hinton Outflow", "Hinton Downstream", "Cuyler and Current Inflow",
"Cuyler and Current Outflow", "Blanchard Upstream", "Blanchard Inflow",
"Blanchard Downstream", "Hinton Upstream", "Hinton Inflow Front Cell",
"Hinton Outflow", "Hinton Downstream", "Cuyler and Dewe Inflow",
"Cuyler and Dewe Outflow", "Blanchard Upstream", "Blanchard Inflow",
"Blanchard Downstream", "Hinton Upstream", "Hinton Inflow Front Cell",
"Hinton Outflow", "Hinton Downstream", "Blanchard Upstream",
"Blanchard Inflow", "Blanchard Downstream"), pH = c(7.84, 6.24,
7.51, 8.25, 8.07, 7.35, 8.11, 8.52, 7.85, 8.01, 7.76, 7.08, 7.51,
8.23, 7.77, 7.09, 7.78, 7.32, 7.04, 6.98, 7.22, 8.27, 7.71, 6.99,
7.88, 8.09, 7.39, 7.96, 8.79, 7.58, 7.22, 6.95, 7.81, 7.41, 7.7,
7.81, 7.68, 7.88, 7.25, 7.82, 7.31, 6.92, 8.05, 6.93, 7.96, 8.03,
8, 7.31, 7.66, 8.05, 7.65, 7.25, 7, 7.9, 6.68, 6.78, 7.86, 8.09,
6.69, 7.85, 7.64, 7.86, 7.57, 7.48, 7.82, 7.72, 7.4, 7.74, 8.04,
7.58, 7.96, 7.72, 7.73, 7.27, 7.56, 7.68, 6.41, 7.1, 7.78, 6.96,
6.65, 7.6, 6.67, 7.99, 7.88, 5.3, 6.97, 7.67, 7.62, 7.37, 7.72,
6.53, 8.03, 8.04, 6.88, 7.06, 8.04, 7.96, 6.07, 7.66), TDS = c(224L,
39L, 165L, 230L, 231L, 25L, 250L, 45L, 42L, 269L, 20L, 44L, 223L,
254L, 170L, 48L, 155L, 20L, 191L, 35L, 188L, 379L, 66L, 99L,
283L, 312L, 62L, 271L, 44L, 47L, 13L, 87L, 219L, 16L, 228L, 204L,
213L, 218L, 19L, 231L, 22L, 197L, 282L, 53L, 193L, 277L, 246L,
27L, 241L, 59L, 61L, 24L, 175L, 210L, 47L, 102L, 201L, 232L,
36L, 211L, 209L, 47L, 182L, 191L, 65L, 166L, 273L, 180L, 50L,
199L, 39L, 49L, 148L, 63L, 153L, 140L, 137L, 159L, 147L, 33L,
410L, 251L, 35L, 268L, 255L, 127L, 150L, 255L, 77L, 150L, 245L,
39L, 243L, 461L, 82L, 140L, 238L, 293L, 47L, 249L), Ec = c(447L,
78L, 330L, 460L, 463L, 51L, 500L, 90L, 85L, 536L, 41L, 88L, 446L,
510L, 340L, 96L, 310L, 40L, 382L, 71L, 377L, 782L, 133L, 199L,
567L, 625L, 124L, 540L, 88L, 93L, 26L, 175L, 438L, 32L, 456L,
408L, 426L, 436L, 38L, 462L, 44L, 395L, 568L, 106L, 386L, 554L,
492L, 55L, 481L, 118L, 123L, 47L, 350L, 421L, 95L, 204L, 401L,
512L, 72L, 422L, 417L, 95L, 364L, 383L, 131L, 338L, 545L, 360L,
99L, 398L, 77L, 100L, 297L, 125L, 308L, 281L, 279L, 318L, 293L,
66L, 819L, 501L, 70L, 536L, 510L, 254L, 300L, 511L, 154L, 301L,
490L, 77L, 487L, 231L, 164L, 280L, 475L, 586L, 95L, 497L), Turbidity..NTU. = c(1.64,
7.25, 5.63, 2.06, 3.08, 5.62, 3.29, 11.1, 3.95, 7.15, 12.9, 7.71,
7.5, 7.1, 21.4, 12.5, 19.6, 3.84, 3.11, 81.7, 20.6, 5.9, 47.2,
63.8, 7.44, 2.92, 9.21, 2.21, 7.31, 6.12, 29.3, 11.6, 6.9, 4.8,
7, 7.11, 9.26, 3.93, 7.28, 4.01, 8.11, 3.59, 6.64, 7.12, 0.25,
5.32, 2.43, 12.5, 6.25, 28.1, 4.95, 4.32, 8.78, 6.42, 27.1, 24.8,
7.21, 3.99, 12.2, 0.7, 16.1, 37, 14, 8.56, 60.6, 7.41, 16.5,
13.2, 98, 43.8, 31.6, 9.42, 12.1, 60.4, 14.9, 34.2, 94, 66.4,
22.1, 55.5, 6.55, 34.4, 4.87, 16.4, 4.07, 644, 25.9, 4.69, 32.9,
27.8, 52.8, 37.9, 53.5, 17.8, 91.7, 26.1, 63, 36.2, 30, 9.07),
TSS..g.L. = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, 8.408,
8.794, 8.794, 8.898, 8.97, 9.67, 8.646, 4.32, 8.52, 8.96,
9.1, 8.85, 8.92, 9.38, 8.9, 8.27, 8.19, 8.59, 8.4, 8.64,
8.5, 8.5, 4.23, 8.59, 8.86, 8.66, 7.98, 8.2, 8.67, 8.55,
8.68, 4.21, 4.48, 4.193, 4.866, 4.28, 4.44, 4.29, 4.24, 4.57,
4.38, 4.21, 4.46, 4.54, 6.2, 5.124, 4.88, 6, 6, 7.2, 6.02,
123, 105, 95, 109, 250, 70.59, 91.95, 117.65, 313.87, 237.7,
98.16, 57.43, 66.797, 248.7, 47.964, 73.333, 484.63, 197.909,
851.33, 4449.04, 2487.91, 127.17, 431.37, 241.76, 177.78,
1107.53, 196.26, 172.73, 233.01, 162.16, 282.21, 482.35,
401.96, 120.97, 821.92, 367.92, 2365.52, 192.1, 299.1, 82
), DOC..mg.L. = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, 16.65,
10.65, 14.73, 14.23, 16.05, 9.114, 21.52, 11.7, 25.6, 16.19,
18.37, 10.04, 11.52, 14.8, 20.12, 18, 12.27, 23.27, 15.02,
25.01, 27.82, 14.2, 9.55, 17.88, 8.045, 16.73, 14.78, 18.15,
18.34, 10.75, 17.67, 13.8, 9.05, 14.97, 23.41, 7.97, 12.75,
12.45, 11.86, 12.63, 7.92, 10.61, 15.7, 7.9, 13.57, 17.73,
19.35, 14.36, 13.08, 28.47, 13.83, 14.95, 13.49, 14.44, 15.2,
17.3, 9.759, 12.16, 14.71, 8.33, 10.02, 10.25, 7.567, 15.89,
31.11, 17.56, 15.45, 17.45, 16.01, 15.73, 24.65, 9.027, 18.34,
26.98, 17.87, 11.88, 16.56, 31.58, 11.81, 15.9, 18.9, 15.79,
21.12, 12.01, 12.1, 16.35, 13.73, 14.11, 15.88, 29.54, 14.21
), Ammonia = c("0.78", "4.32", "0.54", "0.78", "0.43", "1.3",
"0.23", "0.24", "0.36", "0.92", "0.43", "0.6", "0.48", "0.31",
"1.8", "3.44", "2.11", "0.34", "0.3", "0.78", "0.23", "2.52",
"1.24", "1.74", "0.48", "0.35", "0.54", "0.43", "3.36", "0.31",
"0.47", "0.21", "0.35", "0.47", "0.13", "0.56", "0.28", "0.44",
"0.47", "0.39", "0.24", "0.2", "1.42", "0.69", "0.39", "1.49",
"0.36", "1.06", "0.84", "1.58", "0.73", "1.3", "0.78", "1.88",
"0.91", "0.32", "1.42", "0.64", "0.37", "0.48", "0.74", "1.76",
"0.29", "0.27", "3", "7.84", "0.68", "0.18", "0.77", "0.54",
"0.85", "1", "1.82", "0.54", "0.18", "0.67", "0.76", "0.66",
"0.89", "1.02", "0.57", "0.95", "0.79", "0.96", "0.37", "0.85",
"0.52", "0.37", "0.64", "0.79", "0.82", "0.96", "2", "0.29",
"1.42", "0.26", "0.65", "1.34", "1.5", "1.32"), Nitrate = c(0.172,
0.161, 0.155, 0.434, 0.405, 0.124, 0.399, 0.131, 0.178, 1.01,
0.138, 0.845, 0.5, 0.6, 0.25, 0.214, 0.403, 0.15, 0.72, 0.54,
0.61, 0.341, 0.44, 0.329, 0.99, 1.01, 1.327, 1.29, 0.985,
1.67, 0.03, 0.276, 0.15, 0.308, 0.605, 0.299, 0.337, 0.755,
0.144, 0.532, 0.091, 0.135, 0.301, 0.052, 0.194, 0.227, 0.388,
0.291, 0.239, 0.132, 0.484, 0.164, 0.139, 0.349, 0.379, 0.094,
0.281, 0.885, 0.077, 0.069, 0.51, 0.062, 0.227, 0.254, 0.001,
0.246, 0.095, 0.34, 0, 0.028, 0.13, 0.299, 0.304, 0.015,
0.171, 0.198, 0.349, 0.222, 0.195, 0.185, 0.379, 0.161, 0.165,
0.161, 0.293, 0.68, 0.125, 0.066, 0.149, 0.77, 0.319, 0.141,
0.255, 0.178, 0.54, 0.12, 0.177, 0.229, 0.073, 0.228), Nitrite = c(0.004,
0.03, 0.013, 0.029, 0.007, 0.016, 0.007, 0.007, 0.009, 0.004,
0.006, 0.005, 0.055, 0.059, 0.009, 0.024, 0.001, 0.007, 0.004,
0.01, 0.01, 0.035, 0.01, 0.066, 0.021, 0.016, 0.015, 0.02,
0.042, 0.011, 0.008, 0.008, 0.018, 0.009, 0.03, 0.008, 0.012,
0.029, 0.008, 0.037, 0.013, 0.008, 0.01, 0.001, 0.019, 0.016,
0.008, 0.021, 0.006, 0.017, 0.017, 0.016, 0.015, 0.006, 0.001,
0.001, 0.007, 0.006, 0.003, 0.002, 0.008, 0.001, 0.008, 0.064,
0.001, 0.009, 0.001, 0.01, 0, 0, 0.002, 0.007, 0.008, 0.05,
0.032, 0.008, 0.027, 0.009, 0.019, 0.008, 0.008, 0.012, 0.027,
0.041, 0.047, 0.036, 0.033, 0.019, 0.04, 0.019, 0.016, 0.004,
0.042, 0.014, 0.84, 0.019, 0.028, 0.015, 0.001, 0.015), Phosphate = c(0.14,
0.23, 0.11, 0.13, 0.01, 0.14, 0.01, 0.15, 0.16, 0.01, 0.22,
0.29, 0.54, 0.16, 0.36, 0.18, 0.39, 0.16, 0.15, 0.81, 1.76,
0.17, 0.47, 0.53, 0.17, 0.79, 0.93, 0.48, 0.34, 0.63, 0.29,
0.22, 0.17, 0.06, 0.11, 0.13, 0.15, 0.13, 0.01, 0.13, 0.19,
0.31, 0.01, 0.18, 0.32, 0.19, 0.12, 0.29, 0.15, 0.3, 0.91,
0.18, 0.72, 0.18, 0.16, 0.22, 0.13, 0.1, 0.77, 0.15, 0.04,
0.1, 0.06, 0.16, 0.05, 0.38, 0.42, 0.35, 0.43, 0.21, 0.51,
0.38, 0.26, 0.18, 0.22, 0.13, 0.32, 0.22, 0.11, 0.27, 0.01,
0.17, 0.01, 0.01, 0.07, 0.32, 0.24, 0.63, 0.25, 3.5, 0.36,
0.84, 0.2, 0.1, 2.2, 0.26, 0.67, 0.33, 0, 0.32), Zinc = c(0.67,
0.82, 0.33, 0.08, 0.35, 0.29, 0.03, 0.09, 0.1, 1.62, 0.24,
0.05, 0.21, 0.59, 0.28, 0.08, 3.92, 0.63, 1.21, 0.04, 0.26,
0.97, 0.11, 0.13, 0.47, 0.18, 0.63, 0.63, 0.33, 0.25, 0.2,
0.23, 0.38, 0.17, 0.11, 0.32, 0.23, 0.19, 0.15, 0.27, 0.55,
0.3, 2.44, 0.31, 0.16, 2.13, 1.02, 0.14, 1.06, 0.15, 1.05,
0.3, 0.21, 0.15, 0.13, 0.26, 0.14, 0.56, 0.4, 0.46, 0.18,
0.4, 0.26, 0.45, 0.01, 0.29, 0.22, 0.45, 0.25, 0.63, 0.26,
0.13, 0.19, 0.7, 0.76, 0.09, 0.73, 0.22, 0.13, 0.23, 0.21,
0.01, 0.04, 0.01, 0.01, 1.27, 0.05, 0.01, 1.01, 0.17, 1.14,
1.09, 0.4, 0.61, 1.72, 0.74, 0.56, 0.6, 0.04, 0.6), Iron = c(0.17,
0.14, 0.09, 0.12, 0.28, 0.06, 0.32, 0.04, 0.17, 0.1, 0.05,
0.04, 0.17, 0.14, 0.17, 0.01, 0.11, 0.06, 0.18, 0.01, 0.17,
0.11, 0.25, 0.53, 0.12, 0.15, 0.33, 0.23, 0.14, 0.07, 0.07,
0.01, 0.13, 0.04, 0.07, 0.07, 0.21, 0.27, 0.14, 0.15, 0.11,
0.05, 0.08, 1, 0.06, 0.53, 0.1, 0.04, 0.09, 0.07, 0.21, 0.04,
0.03, 0.28, 0.28, 0.37, 0.25, 0.24, 0.05, 0.11, 0.09, 0.31,
0.06, 0.22, 0.16, 0.08, 0.07, 0.42, 0.11, 0.01, 0.28, 0.01,
0.11, 0.27, 0.08, 0.06, 0.17, 0.14, 0.17, 0.01, 0.02, 0.01,
0.01, 0.01, 0.01, 0.37, 0.24, 0.01, 0.28, 0.06, 0.22, 0.12,
0.09, 0.11, 0.28, 0.21, 0.09, 0.19, 0.04, 0.18), Chloride = c(16,
14, 3.4, 7.4, 11.5, 0, 8.3, 2.7, 22, 16, 30, 21, 8.4, 16,
15.5, 14, 16, 8, 15, 9.3, 19.5, 13, 11.5, 11.5, 17.5, 10.5,
4.7, 16, 13, 1.4, 29, 18, 9.1, 21, 4.9, 25, 5.7, 5.9, 10,
6.2, 17.5, 7.2, 34, 0.01, 7.7, 14, 6.9, 2, 11, 13.5, 10.5,
25, 18, 5.6, 11, 3.3, 5.7, 6.3, 5.3, 8.2, 21, 11.5, 23, 7.8,
0.7, 4.7, 21.5, 8.2, 0.01, 10.5, 15, 21, 20.5, 4.4, 4.5,
3.3, 0.01, 6.2, 3.4, 10, 18, 0.5, 0, 5, 2.8, 0, 3.2, 3.7,
0, 0, 13.5, 10.5, 9.2, 15, 18.5, 12.5, 11.5, 11, 12.5, 11
)), row.names = c(NA, 100L), class = "data.frame")