Hello,
I am doing cluster analyzes (ordination + cluster).
Given the "aspect" of my data, I standardized by indicating proportions then I transformed them with asin(sqrt).
First of all to create my dissimilarity matrix I chose the Bray-Curtis method
For my dendrogram I decided to do the UPGMA method and Complete linkage.
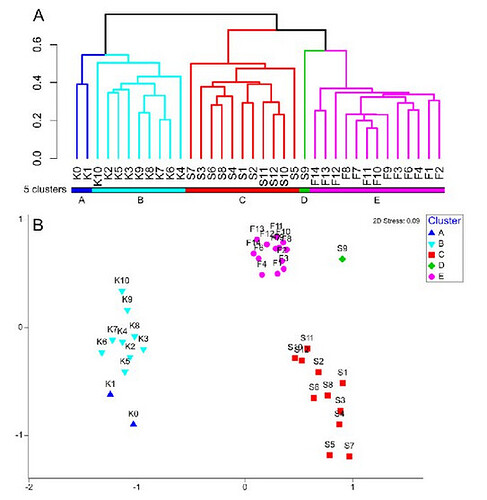
That's my little problem. I managed to make my dendrogram by determining 6 clusters, which I was able to color on the latter to distinguish well.
Now to do my nMDS, I would like the colors of my sites that have been assigned in the dendrogram
dfZ.asin.complete <- hclust(dist.mat,method="complete")
dend <- as.dendrogram(dfZ.asin.complete)
dend <- dend %>%
color_branches(k=6) %>%
set("branches_k_color", c("red", "blue", "green", "black", "orange","purple"), k=6) %>%
set("labels_col", value= c("red", "blue", "green", "black", "orange","purple"), k=6) %>%
set("leaves_pch",19) %>%
set("leaves_col",c("black"))
par(xpd=TRUE, mar=c(8,4,4,3))
plot(dend, main = "Bray-asin(sqrt) dendrogram", ylab = "Height", horiz = T)
How can I make my nMDS analysis to be abble to have something like that :
Thanks !
Hersh