Hi Community
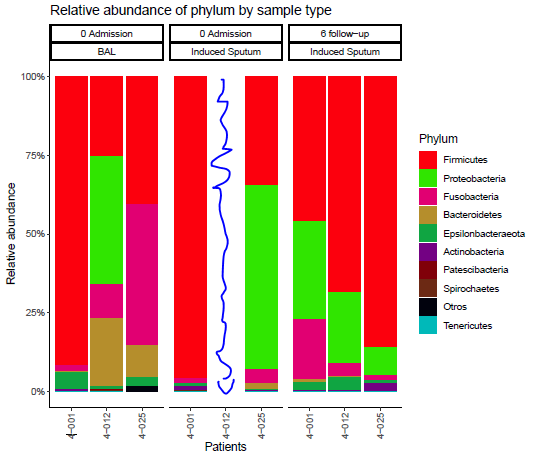
I want to eliminate the patient (4-012) with this interaction (0 Admission with Induced Sputum) on the x-axis, since it does not exist in the database. Only is this value not in the interaction, because in the other events it does exist.
d1 <- d %>%
group_by(Phylum) %>%
mutate(
mean = mean(`Relative abundance`)
) %>%
ungroup() %>%
arrange(desc(mean)) %>%
mutate(
Phylum = factor(Phylum, levels = unique(Phylum)) # maybe this es the problem
)
ggplot(d1) +
geom_bar(aes(x=Patient, y=`Relative abundance`,fill=Phylum), position="fill", stat="identity") + scale_x_discrete("Patients") +
scale_y_continuous("Relative abundance",labels=scales::percent) +
labs(title = "Relative abundance of phylum by sample type") +
theme_classic() +
scale_fill_manual(values=c ("#FC000D", "#30E500", "#E10072", "#B58E2C","#10A542","#730183","#800009",
"#6C2914", "#01020D","#00B9B9"))+
facet_grid(~`Time point`+ `Sample type`)+
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
d<-structure(list(Group = c("HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP",
"HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP",
"HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP",
"HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP",
"HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP",
"HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP",
"HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP",
"HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP",
"HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP",
"HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP",
"HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP",
"HIV+CAP", "HIV+CAP", "HIV+CAP", "HIV+CAP"), Patient = c("4-001",
"4-001", "4-001", "4-012", "4-012", "4-025", "4-025", "4-025",
"4-001", "4-001", "4-001", "4-012", "4-012", "4-025", "4-025",
"4-025", "4-001", "4-001", "4-001", "4-012", "4-012", "4-025",
"4-025", "4-025", "4-001", "4-001", "4-001", "4-012", "4-012",
"4-025", "4-025", "4-025", "4-001", "4-001", "4-001", "4-012",
"4-012", "4-025", "4-025", "4-025", "4-001", "4-001", "4-001",
"4-012", "4-012", "4-025", "4-025", "4-001", "4-012", "4-012",
"4-025", "4-025", "4-001", "4-001", "4-001", "4-012", "4-012",
"4-025", "4-001", "4-025", "4-025", "4-001", "4-001", "4-001",
"4-012", "4-012", "4-025", "4-025"), `Time point` = c("0 Admission",
"0 Admission", "6 follow-up", "0 Admission", "6 follow-up", "0 Admission",
"6 follow-up", "0 Admission", "0 Admission", "0 Admission", "6 follow-up",
"0 Admission", "6 follow-up", "0 Admission", "6 follow-up", "0 Admission",
"0 Admission", "0 Admission", "6 follow-up", "0 Admission", "6 follow-up",
"0 Admission", "6 follow-up", "0 Admission", "0 Admission", "0 Admission",
"6 follow-up", "0 Admission", "6 follow-up", "0 Admission", "6 follow-up",
"0 Admission", "0 Admission", "0 Admission", "6 follow-up", "0 Admission",
"6 follow-up", "0 Admission", "6 follow-up", "0 Admission", "0 Admission",
"0 Admission", "6 follow-up", "0 Admission", "6 follow-up", "6 follow-up",
"0 Admission", "0 Admission", "0 Admission", "6 follow-up", "6 follow-up",
"0 Admission", "0 Admission", "0 Admission", "6 follow-up", "0 Admission",
"6 follow-up", "0 Admission", "6 follow-up", "0 Admission", "0 Admission",
"0 Admission", "0 Admission", "6 follow-up", "0 Admission", "6 follow-up",
"6 follow-up", "0 Admission"), `Sample type` = c("BAL", "Induced Sputum",
"Induced Sputum", "BAL", "Induced Sputum", "BAL", "Induced Sputum",
"Induced Sputum", "BAL", "Induced Sputum", "Induced Sputum",
"BAL", "Induced Sputum", "BAL", "Induced Sputum", "Induced Sputum",
"BAL", "Induced Sputum", "Induced Sputum", "BAL", "Induced Sputum",
"BAL", "Induced Sputum", "Induced Sputum", "BAL", "Induced Sputum",
"Induced Sputum", "BAL", "Induced Sputum", "BAL", "Induced Sputum",
"Induced Sputum", "BAL", "Induced Sputum", "Induced Sputum",
"BAL", "Induced Sputum", "BAL", "Induced Sputum", "Induced Sputum",
"BAL", "Induced Sputum", "Induced Sputum", "BAL", "Induced Sputum",
"Induced Sputum", "Induced Sputum", "BAL", "BAL", "Induced Sputum",
"Induced Sputum", "Induced Sputum", "BAL", "Induced Sputum",
"Induced Sputum", "BAL", "Induced Sputum", "Induced Sputum",
"Induced Sputum", "BAL", "Induced Sputum", "BAL", "Induced Sputum",
"Induced Sputum", "BAL", "Induced Sputum", "Induced Sputum",
"Induced Sputum"), Phylum = structure(c(1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L, 7L, 8L, 8L,
8L, 8L, 8L, 8L, 9L, 9L, 9L, 10L, 10L, 10L, 10L, 10L, 10L, 10L
), .Label = c("Firmicutes", "Proteobacteria", "Fusobacteria",
"Bacteroidetes", "Epsilonbacteraeota", "Actinobacteria", "Patescibacteria",
"Spirochaetes", "Otros", "Tenericutes"), class = "factor"), Counting = c(33540,
29550, 48681, 30346, 59661, 2754, 61297, 23253, 36, 20, 32809,
48562, 19447, 2, 6316, 39114, 669, 440, 20114, 12788, 3566, 3036,
1013, 2973, 100, 46, 1222, 25580, 366, 693, 104, 1232, 1936,
279, 2695, 1173, 3541, 189, 604, 294, 234, 454, 203, 93, 323,
1770, 82, 5, 413, 4, 57, 26, 15, 43, 11, 270, 16, 4, 44, 104,
14, 17, 11, 14, 119, 18, 5, 7), `Relative abundance` = c(0.943497337025182,
0.831256598362973, 1.36942140321177, 0.853648485073529, 1.67829441336492,
0.0774714271367725, 1.72431592926752, 0.654118770955473, 0.00101269839394474,
0.000562610218858188, 0.922933933525915, 1.36607387240957, 0.547054046306759,
5.62610218858188e-05, 0.177672307115416, 1.10029680502096, 0.0188193118208064,
0.0123774248148801, 0.56581709710568, 0.359732973937926, 0.100313402022415,
0.085404231222673, 0.0284962075851672, 0.0836320090332697, 0.00281305109429094,
0.00129400350337383, 0.0343754843722353, 0.719578469919623, 0.0102957670051048,
0.0194944440834362, 0.00292557313806258, 0.0346567894816644,
0.0544606691854726, 0.00784841255307172, 0.0758117269911409,
0.0329970893360327, 0.0996101392488422, 0.00531666656820988,
0.0169908286095173, 0.00827037021721537, 0.0065825395606408,
0.0127712519680809, 0.00571049372141061, 0.00261613751769058,
0.00908615503455974, 0.0497910043689497, 0.00230670189731857,
0.000140652554714547, 0.0116179010194216, 0.000112522043771638,
0.00160343912374584, 0.000731393284515645, 0.000421957664143641,
0.0012096119705451, 0.000309435620372004, 0.00759523795458554,
0.000450088175086551, 0.000112522043771638, 0.00123774248148801,
0.00292557313806258, 0.000393827153200732, 0.00047821868602946,
0.000309435620372004, 0.000393827153200732, 0.00334753080220622,
0.000506349196972369, 0.000140652554714547, 0.000196913576600366
), mean = c(1.01650304554977, 1.01650304554977, 1.01650304554977,
1.01650304554977, 1.01650304554977, 1.01650304554977, 1.01650304554977,
1.01650304554977, 0.514457816751663, 0.514457816751663, 0.514457816751663,
0.514457816751663, 0.514457816751663, 0.514457816751663, 0.514457816751663,
0.514457816751663, 0.156824082192852, 0.156824082192852, 0.156824082192852,
0.156824082192852, 0.156824082192852, 0.156824082192852, 0.156824082192852,
0.156824082192852, 0.103179197824724, 0.103179197824724, 0.103179197824724,
0.103179197824724, 0.103179197824724, 0.103179197824724, 0.103179197824724,
0.103179197824724, 0.0376632378386878, 0.0376632378386878, 0.0376632378386878,
0.0376632378386878, 0.0376632378386878, 0.0376632378386878, 0.0376632378386878,
0.0376632378386878, 0.012694897724093, 0.012694897724093, 0.012694897724093,
0.012694897724093, 0.012694897724093, 0.012694897724093, 0.012694897724093,
0.00284118160523385, 0.00284118160523385, 0.00284118160523385,
0.00284118160523385, 0.00284118160523385, 0.00168314223808408,
0.00168314223808408, 0.00168314223808408, 0.00168314223808408,
0.00168314223808408, 0.00168314223808408, 0.00151904759091711,
0.00151904759091711, 0.00151904759091711, 0.000767561084299385,
0.000767561084299385, 0.000767561084299385, 0.000767561084299385,
0.000767561084299385, 0.000767561084299385, 0.000767561084299385
)), row.names = c(NA, -68L), class = c("tbl_df", "tbl", "data.frame"
))
Thanks
